Abstract
Developmental dysplasia of the hip (DDH) is a painful orthopedic malformation diagnosed at birth in 1–3% of all newborns. Left untreated, DDH can lead to significant morbidity including long-term disability. Currently the condition is clinically diagnosed using 2-D ultrasound (US) imaging acquired between 0 and 6 mo of age. DDH metrics are manually extracted by highly trained radiologists through manual measurements of relevant anatomy from the 2-D US data, which remains a time-consuming and highly error-prone process. Recently, it was shown that combining 3-D US imaging with deep learning-based automated diagnostic tools may significantly improve accuracy and reduce variability in measuring DDH metrics. However, the robustness of current techniques remains insufficient for reliable deployment into real-life clinical workflows. In this work, we first present a quantitative robustness evaluation of the state of the art in bone segmentation from 3-D US and demonstrate examples of failed or implausible segmentations with convolutional neural network and vision transformer models under common data variations, e.g., small changes in image resolution or anatomical field of view from those encountered in the training data. Second, we propose a 3-D extension of SegFormer architecture, a lightweight transformer-based model with hierarchically structured encoders producing multi-scale features, which we show to concurrently improve accuracy and robustness. Quantitative results on clinical data from pediatric patients in the test set showed up to 0.9% improvement in Dice score and up to a 3% smaller Hausdorff distance 95% compared with state of the art when unseen variations in anatomical structures and data resolutions were introduced.
Introduction
Developmental dysplasia of the hip (DDH) is a painful and potentially disabling malformation of that joint affecting approximately 3% of newborns [ ]. DDH can be treated non-invasively if diagnosed early, but missed diagnoses can lead to deteriorating joint health requiring corrective surgeries or early hip replacement.
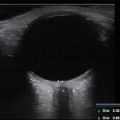
2-D ultrasound (US) scanning is the standard imaging technique for screening DDH in infants under 6 mo old. Cross-sectional views of the hip are manually selected and analyzed by expert radiologists to measure DDH metrics from the geometric properties of the ilium and acetabulum on the pelvis, and of the femoral head [ , ]. DDH metrics acquired using 2-D US have been shown to be highly sensitive to probe positioning, resulting in high intra- and inter-user variability, potentially leading to incorrect diagnoses [ , ]. In comparison, volumetric (3-D) US of the hip followed by automatic extraction of 3-D versions of DDH metrics has been shown to reduce variability by over 70% compared with 2-D US [ ]. These automated approaches implement processes for identifying key structures of the hip joint, including the ilium and acetabulum on the pelvis, along with the femoral head [ , ]. Initial automated approaches used classical statistical or machine learning-based approaches, but more recent methods are based on artificial intelligence/deep learning (AI/DL) approaches.
Early automatic or semi-automatic DDH metric extraction techniques did not reliably generalize to new data [ , ]. Subsequently, 3-D convolutional neural network (CNN)-based architectures, particularly those architectures based on the UNet [ ] architecture, were introduced for 3-D US segmentation [ , ] and showed improvements in segmentation performance. Kannan et al. [ ] used Bayesian techniques to add an uncertainty-based loss term that boosted overall segmentation performance and robustness.
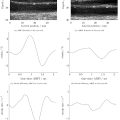
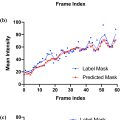
However, despite reports of improved diagnostic repeatability using UNet-based architectures, our group has found a number of situations where acquired US volumes with apparently modest changes from the training data resulted either in implausible, obviously incorrect or failed segmentations and DDH metrics (see Fig. 1 ). Such data variations could arise either from differences in US settings (depth of focus, frequency, etc.) or clinical considerations (age of patients, US probe positioning, patient size, etc.).

A possible reason for the “brittleness” of networks based on CNN architectures is that their restricted receptive fields in the initial layers limit processing of information to localized areas. This can bias the networks’ focus to local features and make it harder to model crucial longer distance relationships between image features.
As an alternative to CNNs, vision transformers (ViTs) use multi-head self-attention layers that grant the network a more global view of the data [ ], which can improve robustness to occlusion, distribution shifts such as stylization to remove textural cues, and to natural corruptions such as noise, blur and pixelation. These characteristics could potentially address the issue of the limited receptive fields of CNNs and help a network generalize better in the presence of anatomical variability typically observed in 3-D US scans of pediatric hips, as has been observed in other medical imaging applications [ ]. For transformers to achieve performance comparable with CNNs, they often need large numbers of both parameters and training data [ ]. To address these concerns, some recent studies have investigated the SegFormer architecture, which is a lightweight transformer-based architecture with hierarchically structured encoders whose multi-scale features are passed into a multi-layer perceptron (MLP)-based decoder [ ].
We therefore hypothesize that a 3-D extension of the SegFormer architecture designed to work directly on 3-D US scans could provide segmentation performance comparable to CNNs, together with improved robustness to data variations common in clinical practice, when trained on comparable amounts of data.
In this study, we quantified the sensitivity of state-of-the-art (SOTA) networks and SegFormer alternative architectures to data variations that might reasonably be encountered in clinical practice. More specifically, we evaluated practical scenarios representative of (i) discrepancies in imaging resolution due to variations in imaging depth settings, (ii) variability in anatomical structure sizes tied to age demographics of the screened population and (iii) variations in the field of view due to changes in US probe positioning, which is heavily reliant on radiologist experience.
Materials and methods
In order to assess the potential improvement in robustness obtainable from alternative transformer-based architectures, we formulated a 3-D extension of the SegFormer architecture and evaluated it against 10 existing DL models on the task of pelvis segmentation from a set of 98 clinical 3-D US scans of newborns from 34 different patients. The 10 DL models evaluated comprised the following: UNet, UNet++ [ ], nnUNet [ ], ResUNet [ ], TransBTS [ ], Attention UNet [ ], Swin UNet [ ], Swin UNetr [ ], UNetr [ ] and PvT [ ].
Extending SegFormer architecture to 3-D
The original SegFormer architecture is a multi-level hierarchical network that learns features at different scales and passes features from all scales into a MLP block [ ]. Figure 2 illustrates our implementation of a 3-D extension to SegFormer, which allows this approach to be applied to volumetric data.

The SegFormer architecture generates a coarse feature map at the output of the MLP decoder at one quarter of the original resolution. We augmented the original implementation by using this coarse feature map and (i) implementing cues from CNN-based encoder-decoder architectures to up-sample the features and (ii) integrating skip connections to pass features from encoder to decoder to provide finer details. Through this approach, we exposed the decoder to high-level features that took into consideration global information along with finer-resolution details passed into the encoder.
Our SegFormer architecture used the up-sampling blocks to produce a volume with the equivalent resolution as the inputs. As the original SegFormer architecture outputs were one quarter of the original resolution, we trained the two convolutional blocks on the right of Figure 2 to up-sample the predictions back to the original resolution. The first block took the predictions from one quarter of the original resolution to one half and the second block up-sampled it back to the resolution equivalent to the inputs.
Self-attention
As described in [ ], self-attention (SA) is a function that maps a set of queries and key value pairs to a tensor. This output tensor is calculated as the dot product between the values and the scaled softmax of the dot product of the queries and keys. Eqn (1) shows how attention is computed where Q is the query, K is the keys and V is the value, and <SPAN role=presentation tabIndex=0 id=MathJax-Element-1-Frame class=MathJax style="POSITION: relative" data-mathml='1dk’>1??√1dk
1 d k
is a scaling factor:
SA(Q,K,V)=softmax(QKTdk)V
The queries, keys and values are generated through fully connected layers connected to the outputs of the initial convolutional blocks. These tensors are then compared with each other through eqn (1) to perform SA.
Extending the attention formulation to 3-D
ViTs split 2-D images into patches and flatten them into tensors [ ]. The complexity of SA is O ( n 2 × d ), where n is the number of patches or sequence length and d is the dimensionality of each element in the sequence. Splitting volumes into cubes results in a significant increase in computational complexity compared with 2-D, as the number of cubes in 3-D is an order of magnitude greater than the number of patches in 2-D. Given our computational resources, passing the entire volume into our SegFormer3D model was computationally intractable due to the number and size of the patches. Thus, given an US volume x ∈ R 3 , we added a feature extractor consisting of convolutional layers that down-sampled the volume from the full resolution to <SPAN role=presentation tabIndex=0 id=MathJax-Element-3-Frame class=MathJax style="POSITION: relative" data-mathml='C×H2×W2×D2′>?×?2×?2×?2C×H2×W2×D2
C × H 2 × W 2 × D 2
, where H, W and D denote the volume dimensions, and C is the number of channels. This is denoted by the blue blocks on the left in Figure 2 .
To further reduce the computational complexity, we reduced the sequence length by deploying an efficient SA block as described by Wang et al. [ ]. As shown in eqn (2 ), this process reduced the sequence length by a ratio R , where S is the sequence to be shortened, N is sequence length, C is the channel dimension and R is the reduction ratio:
S^=Reshape(NR,C·R)(S)S=Linear(C·R,C)(S^)
The original ViT uses positional encoding (PE) to retain positional information after splitting an image into patches. The PE resolution is fixed at training time, which means at test time the PE needs to be adjusted and shows a reduction in performance [ ]. The SegFormer architecture drops the usage of PE and incorporates the Mix Feed Forward Network (Mix-FFN) module, which uses depth-wise convolutional layers to introduce local information through zero padding, as proved by Islam et al. [ ]. Experiments shown by Xie et al. [ ] show that these convolutional layers are sufficient to provide PEs and show that the Mix-FFN-based architecture outperforms PE-based architectures ( eqn [3]) :
xconv=Conv3x3x3(MLP(xin))xout=MLP(GELU(xconv))+xin
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree