and Gary D. Luker1, 2, 3
(1)
Center for Molecular Imaging, Department of Radiology, University of Michigan, Ann Arbor, MI, USA
(2)
Departments of Biomedical Engineering, University of Michigan, Ann Arbor, MI, USA
(3)
Department of Microbiology and Immunology, University of Michigan, Ann Arbor, MI, USA
Abstract
Ligand binding to cell surface receptors activates signaling pathways in normal and pathologic conditions, and internalized ligand–receptor complexes may continue to signal from endosomes. Accessibility of cell surface receptors and the central function of ligand–receptor binding in signal transduction make ligand binding a prime target for therapeutic agents. We describe a Gaussia luciferase complementation method for imaging ligand–receptor binding in cell-based assays and living mice. While we illustrate this imaging method for chemokine ligand CXCL12 and its receptors CXCR4 and CXCR7, this imaging strategy can be generalized to a large number of ligand–receptor interactions.
Key words
Molecular imagingOptical imagingSplit luciferaseBioluminescenceProtein complementation assayPCA1 Introduction
Protein fragment complementation assays based on luciferase enzymes, referred to as luciferase complementation or split luciferase assays, provide a powerful strategy to quantify protein–protein interactions in formats ranging from cell lysates to intact mice. Luciferase complementation entails fusing inactive amino (N)-terminal and carboxy (C)-terminal enzyme fragments to two different proteins of interest. Interactions between proteins of interest bring N- and C-terminal luciferase fragments together to reconstitute an active enzyme, producing bioluminescence as a quantitative measure of interactions between target proteins. Dissociation of proteins of interest also separates fused N-terminal and C-terminal luciferase fragments and reduces bioluminescence, allowing dynamic changes in protein–protein interactions to be quantified in real time. These assays may be used to quantify regulation of protein–protein complexes in response to signaling events, chemical probes, or drugs. When luciferase complementation reporters are expressed stably in mammalian cell lines, the same reporter cells can transition directly from intact cells to animal models, providing a facile approach for development and preclinical testing of candidate drugs.
Monitoring ligand–receptor binding places unique demands on a luciferase complementation assay system. These requirements include that the enzyme functions in the extracellular space and minimizes steric constraints imposed by fusing enzyme fragments to a ligand and receptor. Of available luciferase complementation assays, Gaussia luciferase (GLuc) best meets parameters for imaging ligand–receptor interactions [1]. GLuc does not require ATP, allowing enzyme activity in the extracellular space, and small size of N-terminal and C-terminal enzyme fragments (≈9–10 kDa) substantially reduces the potential to alter functions of fusion proteins. GLuc complementation is fully reversible, so ligand binding to a receptor and subsequent dissociation can be monitored in real time. We have used GLuc complementation to quantify binding of chemokine CXCL12 to its receptors CXCR4 and CXCR7 in intact cells and a mouse model of human breast cancer [2]. More generally, the GLuc complementation system is applicable to any ligand–receptor pair that can be modified to express N-terminal and C-terminal fragments of this enzyme.
2 Materials
2.1 Molecular Biology
1.
DNA encoding open reading frames for desired interacting proteins.
2.
Full-length Gaussia luciferase (GLuc) plasmid (New England Biolabs) or plasmids with NGLuc (amino acids 1–93) and CGLuc (amino acids 94–169) fragments.
3.
Expression vectors with constitutive promoters for expression in mammalian cells.
4.
Expression vector and packaging constructs for producing lentiviral vectors (optional).
5.
Enzymes, buffers, and equipment for PCR, restriction digests of DNA, and ligations
2.2 Cell Culture
1.
HEK-293 T cells or other cell line that can be transfected readily.
2.
Desired cell line(s) for biologic question of interest.
3.
General supplies for cell culture, including media, plasticware, and incubators.
2.3 Cell Imaging
1.
96-Well plates with black sides, clear bottom, and lid for tissue culture.
2.
Multichannel pipets for volumes from 1 to 200 μl.
3.
Sterile pipette tips with low adherence coating adherence.
4.
Sterile 1× phosphate-buffered saline (PBS) solution.
5.
Stock solution of coelenterazine (Promega or other vendor) 1 mg/ml in methanol, stored in tightly closed container at −20 °C (Gaussia luciferase substrate).
6.
Acid wash solution: 0.2 M acetic acid, 0.5 M NaCl (optional) (see Note 1).
7.
Bioluminescence imaging system with high sensitivity and software for data quantification and analysis (IVIS, Perkin-Elmer; or similar system).
2.4 Animal Imaging
1.
Appropriate mouse strain for desired experimental system, such as immunocompromised mice (nude, SCID, or NSG) for human tumor xenografts.
2.
Small animal shaver such as Wahl compact cordless trimmer (optional).
3.
Depilatory solution such as Nair (optional).
4.
10 mg/ml coelenterazine stock in acidified methanol, store in tightly sealed container at−20 ºC (see Note 2).
5.
Sterile solution 40 % DMSO in PBS for diluting coelenterazine immediately before injection and imaging.
6.
28–30 gauge insulin syringe for intravenous tail vein injection in mice.
7.
Restraint device for tail vein injections (Braintree Scientific Tail Vein Injection Platform or other similar device) (optional).
8.
Bioluminescence imaging instrument with isoflurane anesthesia (IVIS or similar instrument as described in Subheading 2.3).
3 Methods
3.1 Construct Gaussia Luciferase Complementation Reporters
1.
Select a ligand and corresponding receptor as interacting proteins and determine positions of NGLuc and CGLuc fusions to these proteins (Fig. 1) (see Note 3).
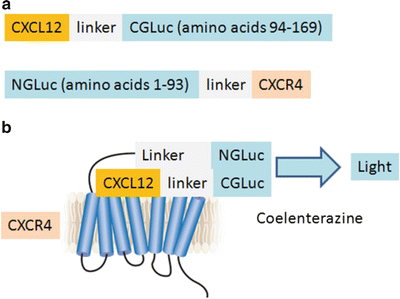

Fig. 1
Schematic diagrams of GLuc complementation constructs and ligand–receptor interaction illustrated for CXCL12 and CXCR4. (a) Complementation reporter constructs with chemokine CXCL12 fused via a linker to CGLuc and NGLuc fused with an intervening linker to seven-transmembrane chemokine receptor CXCR4. These positions of NGLuc and CGLuc enzyme fragments allow complementation to occur in both extracellular and intracellular compartments. We also tested constructs with CXCL12 fused to NGLuc and CGLuc fused to CXCR4, but these reporters produced less bioluminescence upon ligand–receptor binding. (b) Binding of CXCL12-CGLuc to NGLuc-CXCR4 reconstitutes GLuc activity to oxidize the substrate coelenterazine and produce bioluminescence (figure adapted from ref. 10)
2.
Design and optimize linkers between GLuc enzyme fragments and respective ligand and receptor pairs. While not required, linkers may limit steric constraints on ligand–receptor binding and folding of GLuc fragments (see Note 4).
3.
Generate fusion proteins for ligand and receptor pairs using appropriate molecular biology procedures. We typically produce all logical orientations of fusions with NGLuc and CGLuc (see Note 5).
4.
Generate relevant control constructs for nonspecific association of NGLuc and CGLuc (see Note 6).