(1)
Department of Neurosurgery, Cancer Center Amsterdam, Neuro-oncology Research Group, VU University Medical Center, Amsterdam, The Netherlands
(2)
Department of Neurosurgery, Cancer Center Amsterdam, VU University Medical Center, Neuro-oncology Research Group, Amsterdam, The Netherlands
(3)
Department of Neurology, Neuroscience Center, Massachusetts General Hospital and Neuroscience Program, Harvard Medical School, Boston, MA, USA
(4)
Department of Radiation Sciences, Oncology, Umeå University, Umeå, Sweden
Abstract
The use of Gaussia luciferase in a multiplex assay can have several advantages over the singleplex method for an experimental setup. Issues such as intersample variability, screening purposes, efficiency, and in vivo applications can be addressed using a multiplex assay. Here we describe a functional reporter multiplex method using Gaussia luciferase fused to epitope tags to identify the different reporters that are expressed. Tag specific antibodies are used to bind and separate the tagged luciferase reporters.
Key words
Gaussia luciferaseOptical imagingBioluminescent imagingMultiplex assayEpitope tagAntibody binding assay1 Introduction
Bioluminescent reporters can be used to noninvasively detect and quantify biological processes in vitro and in vivo [1]. By engineering the reporter gene under the control of genetic promoters or other gene regulatory elements, the resulting reporter protein reflects the regulation of these elements. Examples of these regulatory elements can be constitutively active promoters to analyze cell survival and proliferation (e.g., CMV promoter) or specific promoters containing transcription factor regulatory elements (TREs) to analyze transcription factor activities [2, 3]. Other examples are 3′-UTR regions to analyze miRNA activities [4] and protease cleavage sites (PCSs) to analyze protease activities [5]. Regularly used bioluminescent reporter genes are Firefly luciferase [6] (Fluc) and Renilla luciferase [7] (Rluc). After translation these luciferases are localized inside the cell’s cytoplasm and ex vivo detection can therefore be a challenge. The enzyme Gaussia luciferase [8] (Gluc) is secreted into the extracellular fluids such as culture medium, animal blood, and urine and can therefore be readily noninvasively detected in these fluids enabling serial sampling over time in a single experiment.
One major disadvantage of luciferase reporters is the difficulty to use a multiplicity of luciferase reporters in combination in a single experiment. This limits the use of luciferase reporters in multiplex assays. Attempts are made to use a range of reporters using modified luciferases, each producing light with a different wavelength in order to discriminate between the different parameters they represent. The downside of this strategy is the need for sophisticated equipment to separate the different signals. Also, overlap in the light spectra might make signal separation a challenge and the number of reporters limited.
Another strategy to analyze multiple reporters in a single assay is to use luciferases that need different enzyme substrates to produce light [9]. A regularly used example is firefly luciferase, which needs d-luciferin as a substrate, in combination with Renilla luciferase, which needs coelenterazine. The disadvantage of this method is the limited number of luciferase substrate combinations currently known.
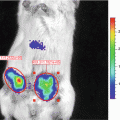
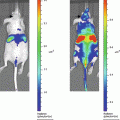
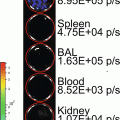
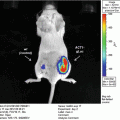
We aimed to develop an easily adaptable method for multiplexing a secreted luciferase reporter in order to perform broad range screening of biologically relevant processes in vitro or in vivo. Therefore, we engineered a range of fusion gene reporters consisting of the Gluc gene, each coupled to a different epitope tag [10] (see Fig. 1). By this method we engineered the ten GlucTag reporters GlucFlag, GlucHis, GlucHA, GlucAcV5, GlucV5 and GlucGlu, GlucMyc, GlucKt3, GlucAu1, and GlucE2. Epitope tags [11] are peptide sequences usually derived from viral or bacterial genes, giving them a high affinity to mammalian antibodies. Coupling the Gluc gene to a range of different epitope tags enables us to design a large number of different GlucTag reporters. Using the corresponding tag specific antibodies to selectively immunobind the different GlucTag makes it possible to separate the different GlucTag reporters and individually quantify them by reading out the light production after addition of substrate [10] (see Fig. 3). As a proof of concept we designed ten different GlucTag based reporters, each coupled to a different epitope tag, under the control of the constitutively active cytomegalovirus (CMV) promoter. After lentiviral transduction [12] we produced ten U87 glioma cell lines, each stably expressing one GlucTag reporter and used these cells to collect GlucTag conditioned cell culture medium or mouse blood to monitor in vitro and in vivo tumor cell growth by measuring GlucTag activity ex situ, after tag specific antibody immunobinding (see Fig. 3). In this chapter the proof of concept of the multiplex Gaussia luciferase-based functional reporter assays in vivo is described [10].


Fig. 1
Overview of the GlucTag construct. We constructed ten GlucTag constructs, each with a different tag: GlucFlag, GlucHis, GlucHA, GlucAcV5, GlucV5 and GlucGlu, GlucMyc, GlucKt3, GlucAu1, and GlucE2
2 Materials
2.1 Construction and Development of GlucTag Reporter Cell Lines
1.
The CSCW-Gluc-IRES-CFP lentiviral vector DNA (see Note 1). This lentiviral vector co-expresses the Gluc bioluminescent reporter and the cerulean fluorescent protein (CFP) control. The internal ribosomal entry site (IRES) allows co-expression of both proteins using the same cytomegalovirus (CMV) promoter. The vector is designated CSCW-Gluc-CFP in this protocol.
2.
AccuPrime Pfx DNA polymerase (Life Technologies, see Note 2). It is provided with a 10× AccuPrime reaction mix and separate MgSO4 (50 mM).
3.
PCR amplification primer oligonucleotides (10 μM, see Note 1):
(a)
Forward: 5′-GCG TGT ACG GTG GGA GGT CT-3′.
(b)
Reverse: 5′-T CTC GAG TAG TCT AGA GTC ACC ACC GGC CCC CTT-3′.
4.
PCR thermocycler.
6.
Heat block.
7.
DNA gel electrophoresis system.
8.
0.5 % agarose (v/w) DNA gel with ethidium bromide (10 μg/ml final concentration). The total volume depends on the DNA gel electrophoresis system used in (item 7). Add for example 0.5 g agarose to 100 ml TAE buffer (item 9) and heat it in a microwave for 2 min at the highest power (~900 W). Make sure that the agarose has melted completely. Cool down the gel to ~65 °C, add ethidium bromide (10 μg/ml final concentration, see Note 4) and swirl to mix. Pour gel in a DNA gel cast with a DNA gel slot comb and let it solidify for 30 min. Remove the DNA gel slot comb carefully.
9.
1× TAE buffer (40 mM Tris, 20 mM acetic acid and 1 mM EDTA in dH2O). Make a 50× TAE stock solution by dissolving 242 g Tris base in dH2O. Add 57.1 ml glacial acetic acid and then add 100 ml 0.5 M EDTA (pH 8.0). Add the volume up to 900 ml with dH2O and set the pH to 8.5 using HCl or NaOH. Bring the total volume to 1,000 ml with distilled H2O and autoclave the TAE buffer for 20 min at 121 °C. Dilute 20 ml 50× TAE stock solution in 980 ml distilled H2O to prepare 1,000 ml 1× TAE buffer.
10.
DNA gel loading dye and a broad range (100 bp to 10,000 bp) DNA gel molecular weight marker. Use the loading dye and the weight marker according to the manufacturer’s guidelines.
11.
DNA gel extraction kit.
12.
T4 DNA ligase.
13.
XL10-Gold ultracompetent cells (Agilent Technologies) for large or ligated DNA.
14.
LB + ampicillin (50 μg/ml) bacteria selection and propagation plates. To make LB, measure 10 g of peptone, 5 g yeast extract, and 10 g NaCl and suspend in 1,000 ml distilled H2O and autoclave the suspension for 20 min at 121 °C. Let the LB cool down to ~65 °C and add ampicillin to a final concentration of 50 μg/ml and briefly swirl. Pour 20 ml of LB + ampicillin per plate (100 mm diameter).
15.
37 °C bacteria incubator.
16.
QIAfilter plasmid kit.
17.
Epitope tag sense and antisense oligonucleotides with appropriate ligation overhangs (see Table 1 and Note 1) in a concentration of 100 μM.
Table 1
Epitope sequences and antibodies used in the immunobinding assays and immunostainings. The epitope tag oligonucleotides are designed to complement each other and to create the proper ligation overhang (XbaI-tag sequence-XhoI). The assay is optimized for use with epitope tag Flag, His, HA, AcV5, V5, and Glu. They show a high binding and assay performance in vitro and in vivo. Epitope tag combinations Myc, Kt3, Au1, and E2 show a low binding and assay performance due to a lower antibody affinity. Conditions for use of these epitope tags or other tags in the assay need to be optimized
Tag | Oligonucleotide sequence: sense, antisense (5′–3′) | Antibody |
Flag | CTAGAGACTACAAAGACCATGACGGTGATTATAAAGATCATGACATCGACTACAAGGATGACGATGACAAGTGAC | Sigma-Aldrich |
TCGAGTCACTTGTCATCGTCATCCTTGTAGTCGATGTCATGATCTTTATAATCACCGTCATGGTCTTTGTAGTCT | F1804 | |
His | CTAGACATCATCACCATCACCACTGAC | Abcam |
TCGAGTCAGTGGTGATGGTGATGATGT | ab81663 | |
HA | CTAGATATCCGTATGATGTGCCGGATTATGCGTGAC | Abcam |
TCGAGTCACGCATAATCCGGCACATCATACGGATAT | ab59076 | |
AcV5 | CTAGAAGCTGGAAGGACGCCAGCGGCTGGAGCTGAC | Abcam |
TCGAGTCAGCTCCAGCCGCTGGCGTCCTTCCAGCTT | ab49581 | |
V5 | CTAGAGGCAAGCCTATCCCTAACCCTCTGCTGGGCCTGGACAGCACCTGAC | Abcam |
TCGAGTCAGGTGCTGTCCAGGCCCAGCAGAGGGTTAGGGATAGGCTTGCCT | ab27671 | |
Glu | CTAGATGCGAGGAAGAGGAATACATGCCTATGGAGTGAC | Abcam |
TCGAGTCACTCCATAGGCATGTATTCCTCTTCCTCGCAT | ab24627 | |
Myc | CTAGAGAACAAAAACTCATCTCAGAAGAGGATCTGTGAC | Sigma-Aldrich |
TCGAGTCACAGATCCTCTTCTGAGATGAGTTTTTGTTCT | M4439 | |
Kt3 | CTAGAAAGCCTCCAACACCTCCACCTGAGCCTGAGACCTGAC | Abcam |
TCGAGTCAGGTCTCAGGCTCAGGTGGAGGTGTTGGAGGCTTT | ab24739 | |
Au1 | CTAGAGACACCTACAGATACATCTGAC | Covance |
TCGAGTCAGATGTATCTGTAGGTGTCT | MMS-130P | |
E2 | CTAGAAGCAGCACCAGCAGCGACTTCAGAGACAGATGAC | Abcam |
TCGAGTCATCTGTCTCTGAAGTCGCTGCTGGTGCTGCTT | ab977 |
18.
10× Oligonucleotide annealing buffer. To prepare a 50 ml stock solution, mix 5 ml 1 M Tris–HCl (pH 7.5, final concentration 100 mM) with 10 ml 5 M NaCl (final concentration 1 M) and 1 ml 0.5 M EDTA (final concentration 10 mM) and add 34 ml ultrapure H2O.
19.
Human embryonic kidney cell line HEK293T.
20.
Human glioblastoma cell line U87.
21.
Dulbecco’s modified Eagle’s medium (DMEM) containing 4.5 g/L glucose, 110 mg/L sodium pyruvate, and 584 mg/L l-glutamine. To make DMEM complete culture medium, add 10 % Fetal bovine serum and penicillin/streptomycin.
22.
Phosphate buffered saline (PBS).
23.
Trypsin (0.5 mg/ml) + EDTA (0.22 mg/ml) in PBS.
24.
A cell culture incubator at 37 °C and with 5 % CO2.
25.
Third-generation lentiviral packaging plasmid pRRE, packaging plasmid pRSV/Rev, and envelope plasmid pMD2.G (Addgene).
26.
3 M Calcium chloride (CaCl2) to make 100 ml of 3 M CaCl2 solution, dissolve 33.3 g CaCl2 × 2H2O in 95 ml ultrapure H2O. Set the pH to 7.2 and fill the solution up to 100 ml. Sterile filter the 3 M CaCl2 solution.
27.
2× HEPES buffered saline (HEBS). Dissolve 8 g NaCl (final concentration 273.7 mM), 0.37 g KCl (final concentration 9.9 mM), 1 g Na2HPO4 (1.4 mM), 1 g dextrose (final concentration 11.1 mM), and 5 g HEPES (42 mM) in 450 ml ultrapure H2O. Set the pH to 7.2 and then fill up the solution to 500 ml with ultrapure H2O. Autoclave the 2× HEBS for 15 min at 121 °C.
2.2 GlucTag-CFP Multiplex Assay Application
2.2.1 GlucTag Immunobinding Assay In Vitro
1.
6-well cell culture plates.
2.
A cell culture incubator at 37 °C and with 5 % CO2.
3.
Wash buffer. Add 0.5 ml Tween-20 (0.05 %) to 1 L of phosphate buffered saline (PBS) to make the PBS + 0.05 % Tween-20 wash buffer solution.
6.
Microplate centrifuge.
7.
Microplate shaker.
8.
Block buffer. Add 0.5 ml Tween-20 (0.05 %) and 10 g bovine serum albumin (BSA, 1 % v/w) in 1 L of phosphate buffered saline (PBS) to make the PBS + 0.05 % Tween-20 + 1 % BSA block buffer solution.
9.
Coelenterazine Gluc substrate. Dilute coelenterazine in methanol (5 mg/ml) to make a stock solution. Before use, dilute the stock solution coelenterazine 1 μl per 1,000 μl PBS + Triton X-100. Incubate the final user solution coelenterazine (5 μg/ml) 30 min at room temperature.
10.
Microplate luminometer.
2.2.2 GlucTag Immunostaining In Vitro
1.
24-well plates.