-space of diffusion signal measurements and fail to take into consideration information redundancy in the 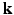 -space. In this paper, we propose a framework, called 6-Dimensional Compressed Sensing diffusion MRI (6D-CS-dMRI), for reconstruction of the diffusion signal and the EAP from data sub-sampled in both 3D
-space. In this paper, we propose a framework, called 6-Dimensional Compressed Sensing diffusion MRI (6D-CS-dMRI), for reconstruction of the diffusion signal and the EAP from data sub-sampled in both 3D 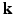 -space and 3D
-space and 3D 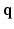 -space. To our knowledge, 6D-CS-dMRI is the first work that applies compressed sensing in the full 6D
-space. To our knowledge, 6D-CS-dMRI is the first work that applies compressed sensing in the full 6D 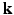 –
–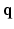 space and reconstructs the diffusion signal in the full continuous
space and reconstructs the diffusion signal in the full continuous 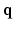 -space and the EAP in continuous displacement space. Experimental results on synthetic and real data demonstrate that, compared with full DSI sampling in
-space and the EAP in continuous displacement space. Experimental results on synthetic and real data demonstrate that, compared with full DSI sampling in 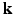 –
–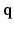 space, 6D-CS-dMRI yields excellent diffusion signal and EAP reconstruction with low root-mean-square error (RMSE) using 11 times less samples (3-fold reduction in
space, 6D-CS-dMRI yields excellent diffusion signal and EAP reconstruction with low root-mean-square error (RMSE) using 11 times less samples (3-fold reduction in 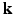 -space and 3.7-fold reduction in
-space and 3.7-fold reduction in 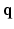 -space).
-space).
1 Introduction
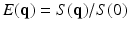 is a continuous function that depends on the diffusion weighting vector
is a continuous function that depends on the diffusion weighting vector 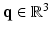 , where
, where 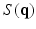 is a diffusion-weighted measurement at
is a diffusion-weighted measurement at 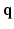 , and S(0) is the measurement without diffusion weighting at
, and S(0) is the measurement without diffusion weighting at 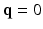 . A central problem in dMRI is to reconstruct the MR signal attenuation
. A central problem in dMRI is to reconstruct the MR signal attenuation 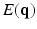 from a limited number of noisy measurements in the
from a limited number of noisy measurements in the 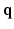 -space and to estimate some meaningful quantities such as the Ensemble Average Propagator (EAP) and the Orientation Distribution Function (ODF). The EAP
-space and to estimate some meaningful quantities such as the Ensemble Average Propagator (EAP) and the Orientation Distribution Function (ODF). The EAP 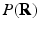 , which is the Fourier transform of
, which is the Fourier transform of 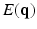 under the narrow pulse assumption [1], fully describes the Probability Density Function (PDF) of water molecule displacements in a voxel. The radial integral of EAP results in the ODF [1], a PDF defined on
under the narrow pulse assumption [1], fully describes the Probability Density Function (PDF) of water molecule displacements in a voxel. The radial integral of EAP results in the ODF [1], a PDF defined on 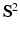 . By assuming a Gaussian EAP, Diffusion Tensor Imaging (DTI) requires only a dozen of measurements for estimating the diffusion tensor for the EAP or the diffusion signal. However, it is well reported that DTI cannot fully characterize complex micro-structure such as crossing fibers [1]. On the other hand, Diffusion Spectrum Imaging (DSI) is a model-free technique for EAP estimation. However, DSI normally requires about 515 signal measurements in
. By assuming a Gaussian EAP, Diffusion Tensor Imaging (DTI) requires only a dozen of measurements for estimating the diffusion tensor for the EAP or the diffusion signal. However, it is well reported that DTI cannot fully characterize complex micro-structure such as crossing fibers [1]. On the other hand, Diffusion Spectrum Imaging (DSI) is a model-free technique for EAP estimation. However, DSI normally requires about 515 signal measurements in 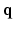 -space, causing a scan time as long as an hour, thus limiting its clinical utility.
-space, causing a scan time as long as an hour, thus limiting its clinical utility.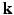 -space CS techniques, such as Sparse MRI [3, 4], have been proposed to reconstruct MR images from a sub-sampled
-space CS techniques, such as Sparse MRI [3, 4], have been proposed to reconstruct MR images from a sub-sampled 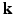 -space, where the sparsity dictionaries are the wavelet basis and the total variation operator. In dMRI, existing techniques mainly focus on applying CS to the
-space, where the sparsity dictionaries are the wavelet basis and the total variation operator. In dMRI, existing techniques mainly focus on applying CS to the 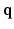 -space [5–7]. References [5, 6, 8] represented diffusion signal and EAP discretely, which suffers from numerical errors in regridding and numerical integration. References [7, 9, 10] represented diffusion signal and EAP continuously, which have closed form expressions of Fourier transform and ODF/EAP calculation. However, this line of work fails to harness information redundancy in the k-space. The correlation of the
-space [5–7]. References [5, 6, 8] represented diffusion signal and EAP discretely, which suffers from numerical errors in regridding and numerical integration. References [7, 9, 10] represented diffusion signal and EAP continuously, which have closed form expressions of Fourier transform and ODF/EAP calculation. However, this line of work fails to harness information redundancy in the k-space. The correlation of the 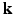 -space and the
-space and the 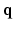 -space can be employed for even greater sub-sampling, thus further reducing scanning while retaining good reconstruction accuracy. To our knowledge, [11, 12] are the only works on signal and ODF reconstruction in joint
-space can be employed for even greater sub-sampling, thus further reducing scanning while retaining good reconstruction accuracy. To our knowledge, [11, 12] are the only works on signal and ODF reconstruction in joint 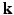 –
–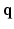 space by using single-shell data (single b value), i.e.,
space by using single-shell data (single b value), i.e., 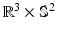 . However, reconstruction of continuous diffusion signal and EAP in whole
. However, reconstruction of continuous diffusion signal and EAP in whole 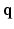 -space
-space 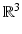 is much more challenging than single shell
is much more challenging than single shell 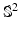 . In this paper, we propose a framework, called 6-Dimensional Compressed Sensing diffusion MRI (6D-CS-dMRI), for reconstruction of the diffusion signal and the EAP from data sub-sampled in both 3D
. In this paper, we propose a framework, called 6-Dimensional Compressed Sensing diffusion MRI (6D-CS-dMRI), for reconstruction of the diffusion signal and the EAP from data sub-sampled in both 3D 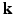 -space and 3D
-space and 3D 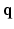 -space. To our knowledge, 6D-CS-dMRI is the first work that applies compressed sensing in the full 6D
-space. To our knowledge, 6D-CS-dMRI is the first work that applies compressed sensing in the full 6D 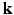 –
–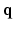 space and reconstructs the diffusion signal in the full continuous
space and reconstructs the diffusion signal in the full continuous 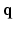 -space and the EAP in full continuous displacement
-space and the EAP in full continuous displacement 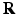 -space. A preliminary abstract of this work was published in [13].
-space. A preliminary abstract of this work was published in [13].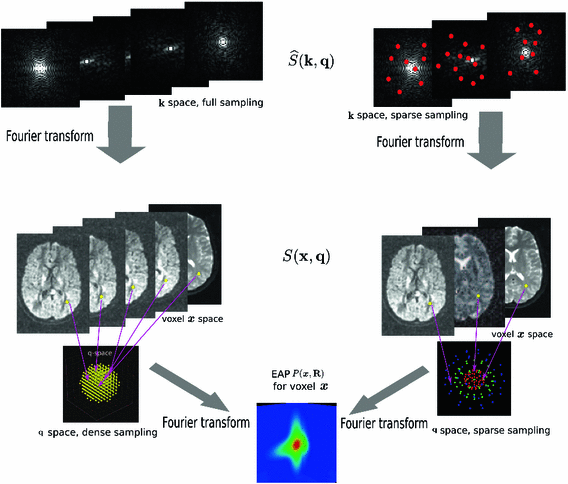
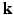 –
–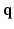 space. Dense sampling (left) and sparse sampling (right) in both
space. Dense sampling (left) and sparse sampling (right) in both 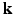 and
and 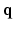 spaces.
spaces.2 Compressed Sensing dMRI in Joint k-q Space
2.1 Sampling and Reconstruction in the 6D Joint 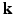 –
–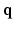 Space
Space
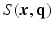 as a complex function in a 6-dimensional (6D) space, i.e. 3D voxel
as a complex function in a 6-dimensional (6D) space, i.e. 3D voxel 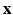 -space and 3D diffusion
-space and 3D diffusion 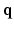 -space, for a fixed
-space, for a fixed 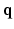 value, the magnitude of
value, the magnitude of 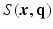 , denoted as
, denoted as 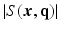 , is a 3D diffusion weighted image volume. Then the
, is a 3D diffusion weighted image volume. Then the 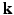 -space measurements
-space measurements 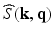 and the EAP are related by [14]
and the EAP are related by [14]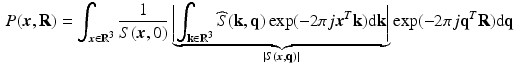
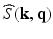 is the 3D Fourier transform of
is the 3D Fourier transform of 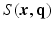 over
over 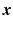 , and
, and 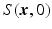 is the image volume with
is the image volume with 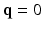 . Two Fourier transforms are involved: the Fourier transform between
. Two Fourier transforms are involved: the Fourier transform between 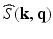 in scanning
in scanning 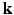 -space and
-space and 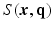 in voxel
in voxel 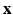 -space for any fixed
-space for any fixed 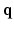 , and the Fourier transform between
, and the Fourier transform between 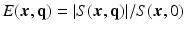 in diffusion
in diffusion 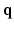 -space and EAP
-space and EAP 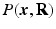 in displacement
in displacement 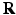 -space for a voxel
-space for a voxel 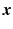 . Instead of dense sampling in
. Instead of dense sampling in 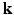 -space and
-space and 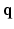 -space, sparse sampling in both spaces can significantly reduce the scanning time. Figure 1 is an overview of the 6D space sampling and reconstruction framework that will be discussed in this paper. The goal is to reconstruct continuous functions
-space, sparse sampling in both spaces can significantly reduce the scanning time. Figure 1 is an overview of the 6D space sampling and reconstruction framework that will be discussed in this paper. The goal is to reconstruct continuous functions 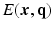 and
and 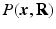 from a small number of samples of
from a small number of samples of 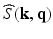 in the joint 6D
in the joint 6D 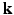 –
–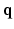 space.
space.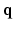 , Sparse MRI can be used to reconstruct the 3D diffusion weighted (DW) images
, Sparse MRI can be used to reconstruct the 3D diffusion weighted (DW) images 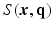 from samples in
from samples in 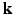 -space [3]. Then all these 3D DW images can be used in a CS-dMRI technique to reconstruct the EAP [6, 7]. This approach separates the estimation into two independent steps. However, the first step fails to take into consideration the diffusion signal in the same voxel across different
-space [3]. Then all these 3D DW images can be used in a CS-dMRI technique to reconstruct the EAP [6, 7]. This approach separates the estimation into two independent steps. However, the first step fails to take into consideration the diffusion signal in the same voxel across different 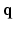 values, and in the second step, information of different voxels in the same DW images is not used.
values, and in the second step, information of different voxels in the same DW images is not used.2.2 6D-CS-dMRI Using Joint Optimization
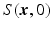 is known or pre-reconstructed by Sparse MRI [3]. The goal here is to estimate
is known or pre-reconstructed by Sparse MRI [3]. The goal here is to estimate 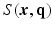 and
and 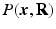 from a number of samples of
from a number of samples of 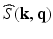 in Eq. (1).
in Eq. (1).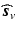 to denote the partial Fourier sample vector of the v-th volume
to denote the partial Fourier sample vector of the v-th volume 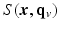 and
and 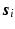 to denote the vector of the diffusion weighted signals
to denote the vector of the diffusion weighted signals 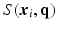 at voxel i with different
at voxel i with different 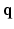 values. We assume that the magnitude of the diffusion signal vector
values. We assume that the magnitude of the diffusion signal vector 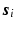 can be sparsely represented by a real basis set
can be sparsely represented by a real basis set 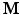 and coefficient vector
and coefficient vector 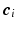 , i.e.
, i.e. 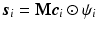 , where
, where 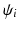 is the complex vector with unit magnitude that contains phase information, and
is the complex vector with unit magnitude that contains phase information, and 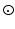 means element-wise multiplication. Then we estimate coefficients
means element-wise multiplication. Then we estimate coefficients 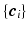 by solving
by solving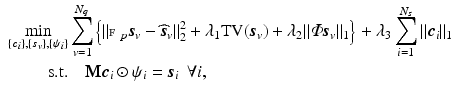
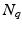 is the number of DW images,
is the number of DW images, 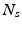 is the number of spatial voxels,
is the number of spatial voxels, 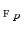 is the partial Fourier transform operator [3],
is the partial Fourier transform operator [3], 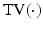 denotes the total variation operator,
denotes the total variation operator, 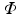 is a chosen wavelet dictionary. Note that
is a chosen wavelet dictionary. Note that 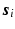 is a complex vector because
is a complex vector because 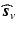 is complex, thus the signal representation
is complex, thus the signal representation 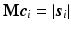 is only applied to the magnitude of
is only applied to the magnitude of 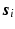 when
when 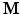 is a real basis set. The first three terms in Eq. (2) originate from Sparse MRI [3]. The sparsity term of
is a real basis set. The first three terms in Eq. (2) originate from Sparse MRI [3]. The sparsity term of 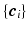 and the equality constraint are from sparse representation in CS-dMRI [6, 7]. Equation (2) is essentially a non-convex optimization problem for variable
and the equality constraint are from sparse representation in CS-dMRI [6, 7]. Equation (2) is essentially a non-convex optimization problem for variable 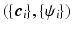 , because of the constraints
, because of the constraints 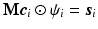 ,
, 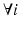 .
.Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree




