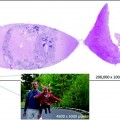
Fig. 3.1
Observations on the same tissue sample from three different pathologists
3.4 Biobanking and Collection Hosting
Translational biomedical research is based on large collections of high-quality samples combined with large sets of well-documented, up-to-date epidemiological, clinical, and/or molecular data from large numbers of patients and controls. Such collections are of the utmost importance in both investigator-driven and company-driven clinical trials. Biobanks are therefore considered essential for the advancement of research and development in the life sciences [11–17]. The term “biobank” is generally defined as an organized collection of human biological material and associated information stored for research purposes. As the term infers, collections of plants, animals, microbes, and other non-human materials could also be labeled biobanks. However, the term is generally reserved for collections of human specimens. Specimen types include blood (in all its forms; e.g., serum, plasma, isolated PBMCs), urine, saliva, skin cells, organ tissue, and other materials taken from the body. Specialized biobanks exist, such as the Network for Pancreatic Organ Donors with Diabetes (nPOD) biobank that houses pancreatic tissues (http://www.jdrfnpod.org) and biobanks that focus on heart valves. The primary task of the biobank is to maintain specimens in good condition for future analysis. For this purpose, biobanks usually have cryogenic storage facilities for the samples, ranging in size from individual refrigerators to warehouses, maintained by institutions such as hospitals, universities, and other non-profit organizations [11–21], but also by pharmaceutical companies (e.g., Astra Zeneca Global Biobank). Disease-oriented biobanks, usually located at a university-based hospital, often have formalin-fixed paraffin-embedded (FFPE) tissue available, next to fresh frozen tissue. Although many collections of human biological materials are present in European countries, these collections often suffer from (geographical) fragmentation, undefined access rules, lack of uniform quality standards, and the absence of any uniform legal and/or ethical framework, thereby hampering international collaboration.
Increasingly, the focus of cancer therapy is shifting toward personalized medicine. For optimal application of targeted molecular drugs, an improved understanding of the underlying molecular mechanisms may help to identify biomarkers that can be used clinically to predict response and establish new treatment options to overcome resistance. For this kind of research, FFPE tissue is the specimen most widely available. Moreover, in contrast with prospective studies, long-term clinical follow-up is an implied characteristic of a biobank. However, the use of high-quality, fresh-frozen biospecimens with appropriate clinical annotation would be preferable. Collections of human biological materials together with associated clinical data are key resources when investigating genetic and environmental factors underlying (multi-factorial) disease, with the aim of improving diagnosis and treatment and ultimately preventing or mitigating disease.
To provide a forum to address the integration of scientific, technical, legal, and ethical issues relevant to repositories of biological and environmental specimens, the International Society of Biological and Environmental Repositories (ISBER, http://www.isber.org) and its chapter covering the region encompassing Europe, the Middle-East, and Africa (ESBB, http://www.esbb.org) were established. These societies aim to create opportunities for sharing ideas and innovations in biobanking, as well as the harmonization of approaches to evolving challenges for biological and environmental repositories. Educational resources and meetings focus on technical issues such as QA and control, regulation, human subject privacy, and confidentiality issues and provide information about sources of equipment and expertise.
However, despite the development of strict protocols for the inclusion of material into biorepositories, specimens may still prove to be of little value for downstream testing. In one large study investigating 1,138 samples from the University of Indiana tissue bank, only 59 % were found to be at least 65 % tumor versus non-neoplastic tissue. Meanwhile, 23 % had a tumor volume that accounted for less than 65 % of the gross specimen, 17 % was entirely negative for tumor, and 1 % was completely necrotic. These findings underscore the importance of instituting adequate measures for histological sample quality control before the release of banked samples for downstream testing [22]. In fact, the availability of an online database of whole slide images for all specimens in a biobank would make it possible for researchers to preselect specimens based on tissue composition.
Finally, a fully digital workflow within pathology departments is within reach [23]. Since whole slide imaging is increasingly used for applications such as education and pathology reviews, it is likely that more laboratories will have access to digital pathology systems which will make whole slide image repositories as add-ons of existing biorepositories more feasible and, in the long term, even mandatory. A whole slide image database accompanying biorepositories should therefore be feasible. This can be taken even one step further by combining WSIs with digital pathology. Image analysis tools can be used to derive objective quantification measures from digital slides. Pattern recognition and visual search tools can be used to classify specimen imagery and identify medically significant regions of digital slides. Incorporating digital pathology into biobank QA procedures, using automated pattern recognition morphometric image analysis to quantify tissue features in digital WSI of tissue sections, can minimize the variability and subjectivity associated with routine pathologic evaluations in biorepositories. Whole slide images and pathologist-reviewed morphometric analyses can be provided to researchers to guide specimen selection [24]. As a specific example, unique spatial–spectral algorithms were developed for applying automated pattern recognition morphometric image analysis to quantify histological tumor and non-tumor tissue areas in biospecimen tissue sections. Measurements were acquired successfully for 75/75 (100 %) lymphomas, 76/77 (98.7 %) osteosarcomas, and 60/70 (85.7 %) melanomas. The percentage of tissue area occupied by tumor varied among patients and tumor types and was distributed around medians of 94 % for lymphomas, 84 % for melanomas, and 39 % for osteosarcomas. Within-patient comparisons from a subset, including multiple individual patient specimens, revealed ≤12 % median coefficient of variation (CV) for lymphomas and melanomas. However, due to phenotypic heterogeneity, the median CV for osteosarcomas was much higher. These data suggest that quantitative image analysis automation can minimize variability associated with routine biorepository pathologic evaluations and enhance biomarker discovery by helping to guide the selection of study-appropriate specimens [25]. An online histopathology system will be an important tool in the valorization of any biobank sample collection. An excellent example is the previously mentioned tissue repository installed by the nPOD-JDRF consortium (www.jdrfnpod.org), an initiative of the Juvenile Diabetes Research Foundation International, a large US patient organization. nPOD biobank houses pancreatic tissues from several hundred organ donors, including patients with diabetes [26]. It has proven to be a highly efficient concept that has attracted researchers and industry from all over the world. The online repository not only provides detailed patient data, including donor demographics and laboratory assays, it also permits digital microscopy access to scanned slides, allowing potential customers to choose and inspect tissue and patient characteristics before ordering tissue sections via their online system [27].
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree