Abstract
Objective
Segmentation of brain sulci in pre-term infants is crucial for monitoring their development. While magnetic resonance imaging has been used for this purpose, cranial ultrasound (cUS) is the primary imaging technique used in clinical practice. Here, we present the first study aiming to automate brain sulci segmentation in pre-term infants using ultrasound images.
Methods
Our study focused on segmentation of the Sylvian fissure in a single cUS plane (C3), although this approach could be extended to other sulci and planes. We evaluated the performance of deep learning models, specifically U-Net and ResU-Net, in automating the segmentation process in two scenarios. First, we conducted cross-validation on images acquired from the same ultrasound machine. Second, we applied fine-tuning techniques to adapt the models to images acquired from different vendors.
Results
The ResU-Net approach achieved Dice and Sensitivity scores of 0.777 and 0.784, respectively, in the cross-validation experiment. When applied to external datasets, results varied based on similarity to the training images. Similar images yielded comparable results, while different images showed a drop in performance. Additionally, this study highlighted the advantages of ResU-Net over U-Net, suggesting that residual connections enhance the model’s ability to learn and represent complex anatomical structures.
Conclusion
This study demonstrated the feasibility of using deep learning models to automatically segment the Sylvian fissure in cUS images. Accurate sonographic characterisation of cerebral sulci can improve the understanding of brain development and aid in identifying infants with different developmental trajectories, potentially impacting later functional outcomes.
Introduction
In the second half of pregnancy, the fetal brain goes through major developmental processes. Cerebral volume increases sixfold from weeks 21 to 38 of gestational age (wGA) and the smooth surface of the fetal brain undergoes a folding process that gives rise to the cerebral sulci and gyri [ ]. This gyrification process takes place in a chronologically predictable manner, and at the end of gestation the distinctive convoluted pattern of the human brain is already perceptible. This folding process shows fast progression from 25 to 35 wGA and starts to decline after term birth [ ]. If we consider that premature birth can happen as early as 22–23 wGA, the importance of the first weeks of life for these infants becomes apparent. Premature infant brain development presents a unique challenge to clinicians and researchers, as its occurrence in extra-uterine conditions may impact brain maturation during a critical period of development. Up to 50% of very pre-term infants will show impaired neurological outcomes, and in a non-neglectable number of them brain damage is not identifiable using conventional neuroimaging studies [ ]. Recent research suggests that brain dysmaturation may be the underlying substrate of some neurological deficits observed in children born pre-term. Information about ways to assess such dysmaturation is largely lacking and mostly limited to magnetic resonance imaging (MRI) studies [ , ].
It has been shown that specific brain sulci and gyri are related to functional development [ , ], and that pre-term infants show alterations in sulcal patterns [ ]. However, spatiotemporal details on pre-term dysmaturation and its functional impact still harbour unresolved questions. The first step in being able to identify an altered folding process is to map the “normal” developing brain. For this reason, research regarding pre-term brain segmentation is becoming of particular interest [ , ]. Most authors use MRI due to its high resolution, although it has inherent limitations for performing sequential studies. In contrast, cerebral ultrasound (cUS) is the most common imaging modality used in neonatal units due to ease of use in a sequential and cost-effective manner, although, as image quality is lower, the segmentation of structures becomes more challenging. However, the possibility of performing sequential examinations makes it of great use for studying the gyrification process in pre-term infants in vivo .
To address this issue, we aimed to generate a weekly atlas of brain maturation using cUS images. In order to accomplish this, segmentation of the main cerebral sulci was required. During the first attempt at atlas generation, a semi-automatic sulci segmentation tool was developed [ ]. This tool employed traditional image-processing methods such as K-means, active contour and thresholding to perform the segmentation task. However, the process of segmenting the different sulci using this application still required significant manual intervention, making it time-consuming and error prone. In addition, recent developments in deep learning algorithms have surpassed the accuracy of traditional image-processing algorithms.
This study presents the first attempt at segmenting brain sulci and gyri in the cUS images of pre-term infants using deep learning-based segmentation algorithms. To test the feasibility of our approach, we focused on a single sulcus, the Sylvian fissure, in a single cUS plane (C3). Figure 1 a shows an example of a cUS image of the C3 plane of a pre-term newborn, while Figure 1 b schematically shows the principal sulcus.

To automate segmentation of the Sylvian fissure, we introduced and analysed different approaches for segmentation, namely the well-known U-Net [ ] and ResU-Net [ ] models. These methods were selected due to their proven effectiveness and robustness in medical image segmentation, particularly in applications requiring high precision at pixel level, including ultrasound images [ ]. U-Net in particular is known for its ability to handle small datasets, which is often a constraint in medical imaging studies, while maintaining reliable performance. Although more recent architectures have been developed, the primary goal of this work was to address a novel problem, i.e. , brain sulci segmentation in ultrasound images, which had not been previously explored. Given the complexity and novelty of this task, we prioritised the use of well-established methods with a track record of success in similar medical imaging challenges to ensure accuracy and interpretability.
These techniques were evaluated quantitatively with a dataset created in this study and validated by two clinical experts in the field. Furthermore, images external to the original dataset were segmented to evaluate generalisation of the models on a dataset different from the original one; in particular, a dataset with images of pre-term newborn brains taken with other US scanners. Fine-tuning techniques were used to segment these new images, providing a comparative study to evaluate the performance of the different configurations used to segment the new images.
Materials and methods
Database
Subjects originated from a prospective, longitudinal cohort of pre-term infants born in two tertiary neonatal units. Pre-term infants born before 32 weeks of gestation between January 2019 and June 2020, without suspicion of genetic disease or major malformations, were eligible. The original database was constructed from 146 infants, and after exclusion of 15 infants with intrauterine growth restriction, 44 from multiple gestation, 4 with relevant brain injury in the first exam and 10 with a C3 plane that did not pass image quality control (the Sylvian fissure was not identifiable in the image), 73 subjects were included for development of the automatic segmentation model. Written consent after oral and written information was provided was obtained from all subjects’ parents. The study was approved by the ethical committee of the participant hospital.
Ultrasound study protocol included a scan within the first 3 days of life, followed by weekly scans until discharge or 40 weeks’ gestational age. For this study, we included all scans performed before 32 weeks of post-menstrual age. Hence, the final number of C3 planes used in this work was 240, as each subject may have had several ultrasound scans. All ultrasound images were obtained by a neonatologist with experience in the field (N.C.) using a My Lab Alpha (Esaote, Genova, Italy) microconvex probe (4–9 MHz), properly anonymised and stored digitally.
External dataset
In addition to the previous dataset, another dataset of images was used to evaluate the generalisation of the models. This new dataset consisted of 36 new C3 planes of pre-term infants born from 24 to 31 wGA. These images were taken using Siemens Acuson S3000 and Canon Applio i700 ultrasound scanners as opposed to the original images, which were taken with an Esaote ultrasound scanner.
Data pre-processing
Before segmentation of the Sylvian fissure, pre-processing of the images was carried out. The original images provided by the clinicians were 800 × 1068 pixels in size, and including labels and tags. During the pre-processing stage the images were cropped to eliminate those tags and labels, allowing the segmentation algorithm to focus only on the neonatal brain. The result of this step produced images of 550 × 868 pixel size.
Ground truth data annotation
In order to both perform training of the deep learning-based segmentation algorithms and evaluate the result of the algorithms, we required manual segmentation of the Sylvian sulcus. To create the manual segmentation masks for the 240 images, we used the MATLAB graphical interface “Image Segmenter” (Image Processing Toolbox, v. R2022b, The MathWorks Inc., MA, USA). In this interface there is a tool called Draw ROIs that allows the creation of regions of interest. All segmentations were validated by two expert clinicians. Figure 2 shows an example of the ultrasound and its corresponding binary mask. Notice that the Sylvian sulcus may appear in both right and left hemispheres (although in some images only one of the two is visible in the image, as we have shown in the results section).

Methodology
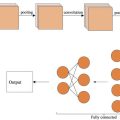
Deep learning architectures. To perform the segmentation task, we explored the use of both U-Net and ResU-Net architectures. The U-Net model has a two-stage U-shaped architecture, with the descending stage called the encoder and the ascending stage called the decoder. In the encoder part, convolutions and max-pooling operations are applied to extract features from the image at the same time as spatial resolution is reduced, allowing for context capturing. In the decoder stage, transposed convolutions and feature concatenation are used to gradually increase the spatial resolution and generate a detailed output and precise localisation of the image mask. The encoder and decoder parts are joined by the bottleneck, with the bottom transition layer compressing information before passing it to the expanding path. Skip connections link corresponding layers in the encoder and decoder paths, ensuring the transfer of fine-grained details for accurate segmentation.
The ResU-Net model is a variant of the U-Net model that uses residual connections to improve gradient propagation during training. These residual blocks are used in both encoder and decoder parts, as well as in the bottleneck. These residual blocks allow information to flow directly through connection hops, avoiding degradation problems during deep network training. Each residual block in ResU-Net consists of two convolutional layers followed by normalisation and activation layers. In addition to the residual blocks, there is another difference between the U-Net and the ResU-Net models implemented in our work. While the U-Net model uses a ReLU activation layer in the final output, the ResU-Net model uses a Softmax activation layer in the channel dimension. This allows generation of a segmentation output with a smoother probability distribution instead of simply a binary mask. In summary, the ResU-Net model allows for more parameters and layers compared with the classical U-Net model. This is due to the introduction of residual blocks, which allow the capture of more complex features and higher level representations.
Figure 3 shows a scheme of the U-Net and ResU-Net models used in this study, detailing the number of levels and the size of each convolutional step.

Fine-tuning. It is well known that deep learning algorithms show a drop in performance when testing images acquired with a different scanner machine to the one(s) used in training. Fine-tuning strategies help to adapt a pre-trained model to the specific characteristics and nuances of the new data domain. Fine-tuning also allows the model to adjust its parameters through continued training on the new data, aligning its representations and predictions with the target dataset. This process enables the model to leverage its prior knowledge while also accommodating the idiosyncrasies of the specific data it will be applied to, ultimately enhancing its performance, generalisation and applicability to the task at hand.
In this work we explored the use of fine-tuning our previous models trained with the in-house data and evaluated their performance using the external dataset acquired with different scanners. For the fine-tuning phase, the full U-Net/ResU-Net models were retrained using a small subset of images from the second dataset. This process involved adjusting the pre-trained model, which was initially trained on a large dataset, with a limited number of new examples. To facilitate gradual and stable adaptation, the learning rate was reduced by a factor of 10 during this phase, where all the network parameters were updated. This approach allowed effective fine-tuning of the model to better align with the new dataset while preserving knowledge gained from the initial training. The combination of using a smaller dataset and a lower learning rate ensured precise adjustments and maintained stability throughout the training process.
Evaluation. The metrics used for evaluating the results of Sylvian fissure segmentation were the dice similarity co-efficient (DSC) and sensitivity. DSC measured the similarity between predicted and ground truth segmentations, indicating how well the model captures the overall shape and overlap. Sensitivity evaluated the proportion of actual positives correctly identified by the model, reflecting its ability to detect true segmented pixels.
Paired t-tests were done to compute the statistical significance of the results. A p -value less than 0.05 was considered significant.
Implementation details
Both models were trained individually. The training process involved running the models for a maximum of 100 epochs, with a patience of 10 epochs. The learning rate for optimisation was set at 0.0001 using the Adam optimiser. To implement the models the PyTorch Lightning library was utilised, with PyTorch serving as the underlying framework. All experiments were conducted on a server equipped with a single NVIDIA GeForce GTX 1080 GPU.
Experimental evaluation
In-house dataset
For evaluating the performance of deep learning methods, we trained the nets using a cross-validation approach and validated them in an independent set. We therefore divided the dataset into two disjointed parts of 204 images for training and 36 for testing. Subsequently, we divided the training data into 6 different folds, each containing 34 images. In the cross-validation strategy, 5 folds where used for training and the remaining fold was used for validation. This process was repeated 6 times, each time using a different part for validation. Finally, the results were averaged to obtain an overall evaluation of the model’s performance. Notice that we ensured that images of the same patient were assigned to the same partition.
To provide an example of the training results, Table 1 presents the results obtained by the ResU-Net model during the six-fold cross-validation of each fold. We can see that the results are similar and stable during each fold, therefore suggesting that the model was not heavily overfitted to any specific subset of the training data, and its predictive ability was relatively uniform across the entire dataset.