(1)
Experimental Therapeutics and Molecular Imaging Laboratory, Department of Neurology, Neuroscience Center, Massachusetts General Hospital, Boston, MA, USA
(2)
Program in Neuroscience, Harvard Medical School, Boston, MA, USA
Abstract
Over the last three decades, imaging has been a thriving field with continuous egression of more reliable and highly sophisticated tools and techniques allowing better understanding of biological processes in living organisms. This field continues to expand and its applications broaden to encompass limitless applications in various biomedical research areas. It is however, of utmost importance to understand the capabilities and limitations of this technique as new challenges and hurdles continue to arise. This chapter describes the general properties of bioluminescence imaging and commonly used reporters while underlining the challenges and limitations with these modalities.
Key words
BioluminescenceImagingPhotoproteinLuxLuciferase1 Introduction
Imaging technologies emerged at the beginning of the twentieth century as a way to complement morphological observations. Molecular imaging (MI) allows a visual (often quantitative) study of molecular, cellular, biochemical, and physiological processes in respect to space and time in a living organism. Advances in molecular and cellular biology, discovery and design of new reporter proteins and molecular probes, as well as the use of transgenic animals have contributed greatly to the expansion of the imaging field.
Imaging systems can be classified under three groups [1, 2]: the energy used to obtain the visual information (X-rays, positrons, photons, or sounds waves); the spatial resolution (macroscopic, mesoscopic, or microscopic); or the type of information acquired (anatomical, physiological, cellular, or molecular). Macroscopic imaging techniques such as computed tomography (CT), magnetic resonance imaging (MRI), and ultrasound are well established in the clinic and provide anatomical and physiological information. Despite the progress in the imaging field, current techniques have not yet been clinically optimized to provide detailed information on specific molecular events (changes in gene expression, activation of certain signaling networks, etc.), especially in the context of the disease (e.g., comparing normal cells versus tumor cells). Hence, new techniques that provide molecular information are currently under development and some are starting to emerge in both preclinical and clinical settings. The most commonly used molecular imaging modalities include positron emission tomography (PET), single-photon-emission CT (SPECT), magnetic resonance (MR), fluorescence-mediated tomography (FMT), laser-scanning confocal microscopy, and bioluminescence imaging (BLI).
Molecular imaging allows a spatiotemporal determination of the molecules of interest as well as monitoring of a specific biological process in living cells or in animals. The applications of this field are numerous and include:
Localization and trafficking of proteins through different cellular compartments using fluorescent reporters fused to the protein of interest.
Monitoring of gene expression using DNA-binding responsive elements acting as promoters to study the expression of certain genes [3].
Visualization of enzymatic activity such as proteases by inserting a small peptide in the middle of the reporter which is recognized and cleaved by the enzyme [4] or LC3 cleavage during autophagy [5, 6].
Molecular imaging, in particular optical imaging (fluorescence or bioluminescence), is also commonly used in drug discovery. Once identified, a molecule hit is validated in animal models using noninvasive imaging techniques to study the drug biodistribution, pharmacokinetics, and potency [12]. In cancer, tumor growth and response to various compounds or gene therapy is commonly monitored using bioluminescence imaging [13, 14]. Unlike histopathological and cytopathological studies, molecular imaging techniques require no chemical fixation or isolation of tissues and organs rendering the study of physiological processes over time in the same biological sample possible. These processes can be determined in their own biological context, while abrogating the need to sacrifice the experimental animals. Data obtained could often be quantified using designed software, which translates the signal into numerical measures.
Bioluminescence imaging offers powerful and versatile tools for monitoring of different biological processes in cultured cells and in living animals. This technique had become indispensable in many molecular biology laboratories, with a diverse and broad range of applications encompassing various biomedical fields and preclinical research areas.
2 Bioluminescent Reporters
Bioluminescence (BL) is the natural production of light often seen in different lower organisms (beetles, bacteria, algae, crustaceans, annelids, mollusks, and coelenterates). Numerous bioluminescent systems exist in nature, many of which have been isolated and studied in laboratories and the biochemical properties of their light emission properly defined. Luminescence is generated through a chemical reaction where the enzyme (luciferase) oxidizes a substrate (luciferin) leading to photon emission. Some luciferases require the presence of cofactors (ATP, Mg2+) for their activity.
Fluorescence is another widely used optical imaging modality that also generates light through a chemical reaction. Unlike BL, this light generation is triggered by an external light source. These two imaging modalities also differ by the signal intensity as well as the signal-to-noise (S/N) ratio. Although fluorescent signals are usually brighter than bioluminescence, the background noise due to autofluorescence is also higher. The high sensitivity of bioluminescence is mostly due to a virtually absent background yielding higher S/N ratios.
Bioluminescent reporters can be divided into two major groups: photoproteins and luciferases. Photoproteins emit light in proportion to the concentration of the protein itself, while in a luciferin-luciferase reaction, photon emission is directly proportional to the amount of luciferin [15].
2.1 Photoproteins
This family encompasses proteins that emit light in proportion to the protein itself and do not require an enzyme (luciferase) [15]. While coelenterate photoproteins have been notorious and widely employed as sensitive reporters for Ca2+ detection, not all photoproteins are Ca2+ sensitive. In fact, other photoproteins, which activity depends on the presence of H2O2, ATP, Mg2+ or superoxide, have been reported [16]. Most studies employ the Ca2+-regulated photoproteins that use coelenterazine as a substrate (aequorin, obelin, phialidin, berovin). Aequorin, from the jellyfish Aequorea victoria is the best-known and widely used photoprotein. It is commonly used to monitor Ca2+ concentrations from a single cell [17]. Like all coelenterate photoproteins, aequorin has an approximate molecular mass of 20 kDa and emits blue light in the presence of Ca2+ [15]. The aequorin protein contains coelenterazine in the central cavity and is capable of binding Ca2+ through its three “EF hand” motifs [18]. Upon binding to Ca2+, the protein undergoes a conformational change, decomposing into apoaequorin while oxidizing coelenterazine into coelenteramide and CO2 with emission of blue light at 469 nm [19] (Fig. 1).
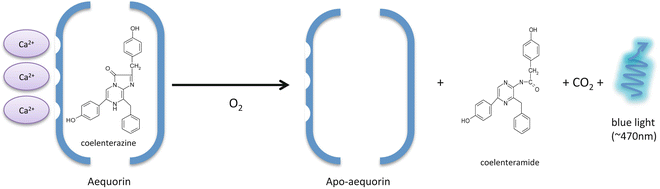

Fig. 1
Schematic representation of the aequorin bioluminescence reaction. When Ca2+ binds to aequorin, conformational change of the apoaequorin protein and oxidation of coelenterazine results in blue light emission
Typically, the recombinant aequorin is expressed in the cell of interest. When coelenterazine is added to the cells, it will passively diffuse and generate bioluminescence light relative to the total intracellular Ca2+ levels available for the reaction. While the high sensitivity of aequorin for Ca2+ make this reporter an ideal Ca2+ sensor, signal acquisition can be a daunting task due to the protein low light quantum yield (number of photons emitted per protein) combined with the low protein stability [20].
2.2 Luciferases (See Table 1)
Table 1
Comparison of different luciferases
Luciferase | Origin | Size (kDa) | Substrate | Cofactors | In vitro sensitivity | In vivo sensitivitya,b | Secreted | Peak emission (nm) |
|---|---|---|---|---|---|---|---|---|
Bacterial luciferase | Various photo-bacterium species | α subunit: 40 β subunit: 37 | FMNH2 | O2, NADPH | + | + | No | 490 |
Firefly | Photinus pyralis | 61 | d-luciferin | O2, ATP, Mg2+ | +++ | ++++ | No | 562 |
Renilla | Renilla reniformis | 36 | Coelenterazine | O2 | ++ | ++ | No | 480 |
Gaussia | Gaussia princeps | 19.9 | Coelenterazine | O2 | ++++ | +++ | Yes | 480 |
Cypridina (Vargula) | Vargula hilgendorfi | 62 | Vargulin | O2 | +++ | +++ | Yes | 460 |
Metridia | Metridia longa | 24 | Coelenterazine | O2 | +++ | + | Yes | 480 |
2.2.1 Bacterial Luciferases (Lux)
This particular group of luciferases uses the reduced riboflavin phosphate (FMNH2) as their substrate, in addition to a long-chain fatty aldehyde and oxygen. FMNH2 is oxidized to emit a blue–green light at 490 nm [21]. The synergistic expression of all five genetic components of the luxCDABE operon produces an autonomous bioluminescence reaction [22]. This property represents a major advantage for lux reporters since light is generated without the need for substrate administration or experimental manipulation. The luxA and luxB genes encode the α- and β-subunit, respectively, which form the heterodimeric luciferase. The luxCDE genes are required for the regeneration of the long-chain fatty aldehyde [22]. The additional components for bacterial luminescence include oxygen and FMNH2, readily available within the cell (Fig. 2). The lux reporters are commonly expressed in bacterial hosts as a means to monitor bacterial growth. Recently, Close et al. described a codon-optimized lux cassette that generates an autonomous bioluminescent system for mammalian expression [23]. This reporter allows whole animal imaging while eliminating the need for substrate administration, a clear advantage over commonly used luciferases such as Firefly, Gaussia, and Renilla (see below). However, the signal intensity remains significantly low as compared to these luciferases. An increase in aldehyde production, which would substantially increase the signal intensity, is hindered by the cytotoxicity of this organic compound [24].
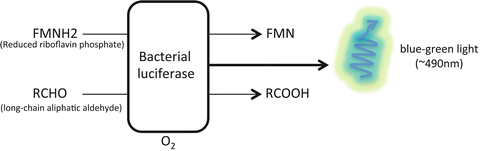

Fig. 2
Schematic representation of the bacterial luciferase reaction. The bacterial luciferase reacts with the reduced riboflavin phosphate in the presence of a long-chain fatty aldehyde to produce a blue–green light
2.2.2 Firefly Luciferase (Fluc)
A monomeric protein (61 kDa) found in the light-emitting organ within the abdomen of the American firefly Photinus pyralis [25]. Fluc is the most studied luciferase due to its high quantum yield (originally thought to be around 88 %, however, a more advanced study using a CCD-spectrometer system showed it to be closer to 41 % [26]). This luciferase requires ATP and Mg2+ as cofactors in combination with its substrate, beetle d-luciferin (a benzothiazole) [1, 27, 28]. Fluc catalyzes a glow-type bioluminescence reaction generating yellow–green light with a peak emission at 562 nm. For Fluc in vivo BLI, d-luciferin injected intraperitoneally (i.p.) or intravenously (i.v.) has a high biodistribution since it can cross the blood- and placental barriers. Maximum light emission is achieved at 10–12 min after i.p. luciferin injection followed by a slow decay over 60 min [29].
2.2.3 Renilla Luciferase (Rluc)
A monomeric protein (36 kDa) from the sea pansy Renilla reniformis. Rluc catalyzes the oxidative decarboxylation of its substrate coelenterazine while emitting blue light with a peak at 480 nm. Like other coelenterates, Rluc generates a flash-type bioluminescence reaction and does not require ATP for activity. A disadvantage of Rluc over Fluc is its low enzymatic turnover and quantum yield (6 %) [30, 31]. Also, the blue emission as well as the poor biodistribution of coelenterazine makes Rluc a less desirable reporter for in vivo imaging [30].
2.2.4 Gaussia Luciferase (Gluc)
A monomeric protein (19.9 kDa) from the marine copepod Gaussia princeps, which uses coelenterazine as a substrate. Gluc is the smallest known luciferase, it is naturally secreted and emits a flash light at a peak of 480 nm with a broad emission spectrum extending to 600 nm [32]. Gluc has several advantages over other luciferases: it is over 2000-fold more sensitive than Fluc or Rluc in reporting from mammalian cells and gives a much stronger bioluminescent signal in vivo [32]; Since it is naturally secreted, it can be detected in the conditioned medium of cells expressing this reporter in cell-based assays, and in the blood or other bodily fluids in small animals [33]. On the other hand and similar to Rluc, the blue light emission and the stability of coelenterazine, makes Gluc less favorable for in vivo BLI.
Other luciferases had been described in the literature. Metridia luciferase, a 24 kDa protein with a peak emission at 480 nm, also utilizes coelenterazine as substrate [34]. Cypridina luciferase (also known as Vargula; Vluc), a 62 kDa protein, has a peak emission at 460 nm and utilizes Cypridina luciferin (vargulin) as substrate [35]. Both of these marine luciferases are naturally secreted and do not require ATP for activity.
2.3 Multiplexing BLI Reporters
Despite the advantages of BL reporters over their fluorescent counterparts, the versatility when it comes to different light-emitting spectra remains a downside. Various mutations in the GFP and other fluorescent proteins resulted in a large palette of colored reporters [36]. Once combined together, these reporter variants can be used to monitor various processes within the same experimental sample. Similar mutation studies, to generate luciferases with various light-emission properties or better stability, are now starting to emerge. Branchini et al. generated a green and red variant of Fluc allowing monitoring of two-different activities simultaneously in the same biological sample [37]. A red-shifted variant of Rluc with a peak emission at 547 nm was shown to be better suited for small animal imaging [38]. Recently, new Gluc variants have been characterized with a glow-type bioluminescence reaction, suited for high-throughput functional screening applications [39, 40]. While the spectral diversity of a given luciferase makes it an attractive tool for a multiplex BL assay, the chemical properties of these reporters are also of high relevance. The type of the BL reaction (flash versus glow) and the different substrate chemistry can be exploited for sequential reading of the different luciferase activities. For example, Rluc (or Gluc) luciferase can be combined with Fluc to measure two-different readouts after addition of coelenterazine and d-luciferin, respectively. Recently, Maguire et al. optimized a triple-imaging platform by combining Fluc, Gluc, and Vluc. Their approach allowed monitoring of gene delivery, tumor size, as well as transcription factor activity within the same animal [41]. Such multiplexing approaches are highly valuable for cell-based high-throughput screening, allowing measurement of various parameters within the same biological sample and gathering a maximum amount of readouts in a time- and cost-efficient fashion.
3 BLI Applications
Bioluminescent organisms often use light-emission properties to interact and communicate with their environment and the surroundings organisms. By reproducing those same BL properties, researchers are able to study similar interactions among cells, genes, and other entities of relevance to the biomedical field.
Several Bioluminescence imaging strategies have been developed to study different cellular and molecular events in living organisms, while providing a spatial and temporal resolution. These imaging paradigms rely on the expression of a foreign protein (luciferase) that is not usually expressed in the cell or the organism of interest. Upon exposure of the luciferase enzyme to the corresponding substrate, light emission can be detected using a luminometer (in cultured cells) or a cooled charge-coupled device (CCD) camera in animal models. Luciferase reporters are commonly used for tracking of cell viability, quantifying gene transfer efficiency, estimating tumor burden or detecting metastatic lesions in animals (Fig. 3). In such studies, the luciferase, expressed under a particular promoter, is introduced to the cell of interest either by plasmid transfection or viral transduction (for stable expression).
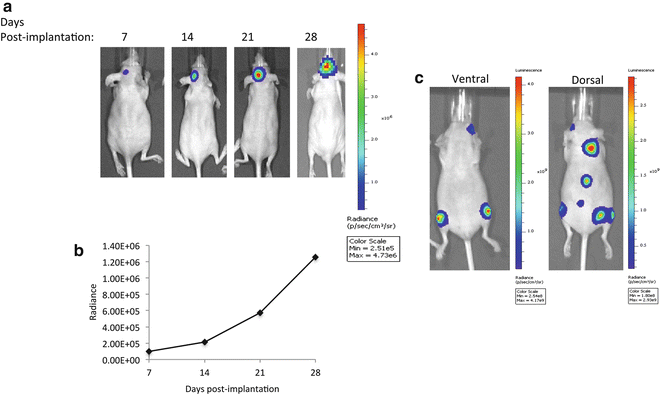

Fig. 3
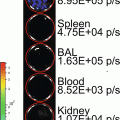
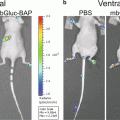
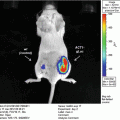
(a, b) Monitoring of intracranial tumor growth using bioluminescence imaging. The human glioblastoma cell line U87-MG-Fluc cells (1 × 105 cells) were stereotactically injected into the left midstriatum of nude mice. Animals were imaged for Fluc at different time points. Representative images from the CCD imaging are shown in (a). Quantification of the BL signal is represented as radiance (photons/s/cm2/sr) in (b). (c) Bioluminescence imaging of a breast cancer metastatic model using a CCD camera. MDA-MB-231-Fluc cells (2 × 105 cells) were implanted intracardially into the animal. Dorsal and ventral images 4 weeks after tumor cells implantation show metastatic lesions in various organs
BLI has also been used for studying different biological and molecular processes such as cell signaling, transcriptional promoters, gene expression, protein–protein interactions, protein conformational changes, enzymatic activities, protein secretion, and visualization of subcellular proteins. For in vitro assays, BL allows the study of different processes at the nuclear, cytoplasmic, and cellular level as well as cell-free based analysis [42]. One major advantage of this imaging technique is its noninvasive nature allowing the study of biological processes in intact living cells or animals. When using a secreted luciferase, it is possible to longitudinally monitor various biological parameters form the same sample by assaying few microliters of the cell culture medium. These reporters also have major advantages for in vivo studies since they can be detected in bodily fluids such as blood, serum, or urine [43]. Due to its high sensitivity, the Gluc secreted reporter has been used to study tumor growth and therapy, viral replication, viability of circulating stem cells as well as metastatic tumors by directly assaying its activity in the blood of experimental animals [33, 44]. While typically in vivo imaging experiments are performed using small animals models (mice, rats), secreted luciferases can be applied for imaging larger animal models. Gluc has already been used as an ex vivo blood reporter to study gene transfer in the lungs of mice and sheep [45]. Mluc and Vluc can also be used as secreted blood reporters. Mluc, however, is inactivated by serum, thus limiting its use as a secreted blood reporter [46, 47].
4 Drawbacks and Limitations
Just like any other imaging modality, BLI has its advantages and its shortcomings. Oftentimes, data interpretation is performed under the assumption that the luciferase activity is directly correlated to the transcriptional activity of the reporter gene, therefore the BL signal is linear in respect to cell number. However, both endogenous and exogenous factors can impact the various components of the luciferase reaction and might lead to erroneous readouts. It is however important to keep in mind that, based on the assay design, luciferin, luciferases, or cofactors such as ATP can be the variable component for the BL reaction [48]. Some of common problems encountered when using luciferase-based reporters are discussed below. These problems can influence the BL signal generated and should be taken into account when designing a BLI experiment.
Signal quantification: It is difficult to standardize in vitro BL assays since the relative light units (RLUs) measured from a luciferase reaction are arbitrary units. The RLU varies largely from one luminometer or photon detector to the other. For example, interlaboratory BL assays using the same luciferase and luciferin but a different luminometer would most likely yield RLUs with great variability. It is very important to keep in mind that BL is rather a semiquantitative method; its sensitivity depends partly on the luminometer when applied in vitro or the CCD camera for in vivo imaging.
Background control: While the high sensitivity of BLI is partly due to its low background noise, it is still recommended to include a proper background control for your assay. For example, when preforming an in vitro BL assay from cellular lysates or cell supernatant, it is important to use the same buffers or media from cells that do not express the luciferase. The serum among other supplements added to cell culture media can impact the BL readings and must be accounted for. Autoluminescence or light emission by substrates such as coelenterazine, in the absence of the enzyme, can also increase the luminescence background and cause variability.
Half–life of the enzyme and stability: these factors can vary greatly between luciferases and can range from few hours to several days. It is important to determine a priori the half-life of your luciferase based on your assay conditions. Both endogenous and exogenous factors can also affect the luciferase stability. In one example, observations made by Czupryna et al. suggested that the Fluc activity is rapidly lost in apoptotic cells due to oxidative stress and particularly hydrogen peroxide which inhibited the BL signal [49]. The different components of the extracellular medium (cell growth medium, blood, urine, etc.) can also significantly affect the enzyme stability notably for secreted luciferases.
Cellular environment: Both secreted and non-secreted luciferases can be subject to different intra or extracellular conditions which could directly affect their activity. Proteolytic degradation of the enzyme, pH, temperature, and H2O2 levels are among many factors that could impact the BL signal. Further, these conditions could indirectly affect the luciferase-expressing cells and impact the proper synthesis, folding, maturation, or secretion of the enzyme.
Oxygen, hypoxia, and oxidative stress: oxygen is a limiting factor for all luciferase reactions. BL assays cannot be conducted under anaerobic conditions, and light emission from hypoxic tissues such as the bulk of a tumor is virtually absent. The lack of oxygen could also indirectly impact the BL reaction by affecting other cofactors needed for certain luciferase reaction (e.g., ATP for Fluc). Moriyama et al. attributed the decrease in Fluc BL signal in hypoxic cells in vitro to an intracellular ATP depletion [50]. In contrast, the Czupryna et al. study attributed this signal decrease in apoptotic cells to hydrogen peroxide [49]. Interestingly, ATP depletion is commonly observed during apoptosis.
Substrate availability: This factor is almost irrelevant for in vitro assays where the substrate and the cofactors needed for the BL reaction are always in excess to the enzyme. In such settings, light emission is directly proportional to the luciferase concentration. However, substrate availability to the luciferase-expressing cells is critical for in vivo imaging. To generate a strong in vivo photon emission, a sufficient amount of substrate should reach the luciferase-expressing cells and (in the case of non-secreted luciferases) should be taken up by these cells. In this setting, the location of the luciferase-expressing cells and their membrane permeability to the substrate, as well as the amount of substrate injected into the animal can impact the BL signal. Larger tumors have higher substrate uptake compared to smaller size, which could lead to inadequately higher signals [42]. Finally, whether the luciferin is metabolized or directly cleared via the kidneys depends on the type of substrate used and on various physiological parameters such as temperature, heart rate, and breathing of the animal. Most frequently, animals are under deep anesthesia during imaging and maintaining similar physiological condition among the different subjects is important to minimize artifacts due to substrate availability.
Luciferin efflux: The luciferase substrate can be actively pumped outside of the cells thus reducing the BL signal. The ATP-binding cassette superfamily of multidrug efflux pumps is notorious for conferring chemoresistance in tumor cells. Two of these efflux pumps, the ABCG2 (BCRP) and ABCB1/Pgp, are substrates for d-luciferin and coelenterazine, respectively [51, 52]. Their expression can significantly reduce the Fluc or Rluc/Gluc signal. Compounds that affect membrane transport proteins could significantly impact the BL signal.
Administration route of the substrate: this factor is very critical for in vivo imaging. 14C-labeled d
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree