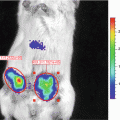
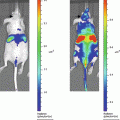
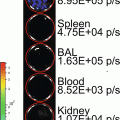
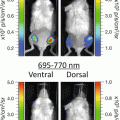
(1)
Department of Imaging and Pathology, Biomedical MRI/MoSAIC, KU Leuven, Leuven, Belgium
(2)
Laboratory of Molecular Cell Biology, Department of Molecular Microbiology, Institute of Botany and Microbiology, VIB, KU Leuven, Leuven, Belgium
Abstract
Fungal biofilms formed on various types of medical implants represent a major problem for hospitalized patients. These biofilms and related infections are usually difficult to treat because of their resistance to the classical antifungal drugs. Animal models are indispensable for investigating host–pathogen interactions and for identifying new antifungal targets related to biofilm development. A limited number of animal models is available that can be used for testing novel antifungal drugs in vivo against C. albicans, one of the most common pathogens causing fungal biofilms. Fungal load in biofilms in these models is traditionally analyzed postmortem, requiring host sacrifice and enumeration of microorganisms from individual biofilms in order to evaluate the amount of colony forming units and the efficacy of antifungal treatment. Bioluminescence imaging (BLI) made compatible with small animal models for in vivo biofilm formation is a valuable noninvasive tool to follow-up biofilm development and its treatment longitudinally, reducing the number of animals needed for such studies. Due to the nondestructive and noninvasive nature of BLI, the imaging procedure can be repeated in the same animal, allowing follow-up of the biofilm growth in vivo without removing the implanted device or detaching the biofilm from its substrate. The method described here introduces BLI of C. albicans biofilm formation in vivo on subcutaneously implanted catheters in mice. One of the main challenges to overcome for BLI of fungi is the hampered intracellular substrate delivery through the fungal cell wall, which is managed by using extracellularly located Gaussia luciferase. Although detecting a quantifiable in vivo BLI signal from biofilms formed on the inside of implanted catheters is challenging, BLI proved to be a practical tool in the study of fungal biofilms. This method describing the use of BLI for in vivo follow-up of device-related fungal biofilm formation has the potential for efficient in vivo screening for interesting genes of the pathogen and the host involved in C. albicans biofilm formation as well as for testing novel antifungal therapies.
Key words
Candida albicans BiofilmBioluminescence imaging Gaussia luciferaseCatheterAnimal modelMouseCoelenterazineInfectious diseasesNoninvasiveIn vivo1 Introduction
In the past decades, the emergence of fungal infections caused by opportunistic pathogenic yeasts (e.g., Candida albicans, Cryptococcus neoformans) and molds (e.g., Aspergillus fumigatus) has increased significantly and has become a common challenge in clinical practice, especially among patients who are immunocompromised or hospitalized with serious underlying diseases [1–3]. Increased numbers of fungal diseases are related to the increasing number of severely ill or immune-compromised patients and in particular to the growing use of implanted medical devices in patients [2, 4, 5]. Candida albicans is an opportunistic human fungal pathogen responsible for the majority of yeast infections in these patients [3, 6–8], causing infections that can range from superficial to systemic and life-threatening candidiasis [7].
Candida biofilms and its characteristics have been mostly elucidated in vitro [9]. However, these models cannot account for numerous host and infection-site variables which are important during the development of biofilm-related infection in humans. Recently, in vivo central venous catheter (CVC) C. albicans biofilm models have been introduced in rats [10], rabbits [11], and mice [12]. These models are technically demanding and of very low throughput. To address this issue, a rat in vivo subcutaneous foreign body infection model has been adapted to study C. albicans biofilms [13, 14]. This subcutaneous catheter rat model system was validated for testing antifungal drugs against mature C. albicans biofilms [15]. Although this model cannot account for factors such as shear stress and nutrient flow, the suitability of the model to evaluate the efficacy of antifungal drugs on biofilm development was shown, allowing more rapid screening under in vivo conditions [15].
In these biofilm models, fungal load is traditionally analyzed post mortem by enumeration of the number of colony forming units attached to the catheter fragments and from individual host organs to determine the extent and kinetics of dissemination during infection. Only one time point per animal can be measured and therefore large groups of animals are needed. These end-point assays do not always provide a full understanding of the dynamic steps in Candida biofilm development, nor do they account for interindividual variation. Noninvasive imaging has the potential to deliver data over the biologically relevant time scale from individual animals in vivo, obviating the need to sacrifice animals at different stages of biofilm development. Among other imaging techniques, bioluminescence imaging (BLI) has emerged as a powerful method to analyze infectious diseases in animal models and microbial viability in particular [16]. Although BLI is well established for imaging different (microbial) cell populations, for yeast cells and other fungal pathogens this is particularly challenging. Because of their cell wall, the diffusion of the necessary substrate for the light producing reaction is limited, hampering detection of a BLI signal from deeper infection sites in vivo [17]. To overcome this problem, a synthetic C. albicans codon-optimized version of the gene for the naturally secreted Gaussia princeps luciferase (gLUC) can be used by tagging gLUC to the C. albicans PGA59 gene, which encodes a glycosylphosphatidylinositol (GPI)-linked cell wall protein [18]. As the luciferase will be present extracellularly, problems with the intracellular delivery of the substrate (coelenterazine, CTZ) are avoided. This expression system has made BLI of superficial C. albicans infections possible [18].
Here we describe a multi-temporal noninvasive imaging assay to follow biofilm formation under in vivo conditions in mice and rats, avoiding the need to sacrifice the animals which results in statistical, ethical, economic, and experimental advantages. The use of in vivo imaging reduced the time, number of animals and costs for the analysis of biofilm models and increased the versatility of the current animal models to the use of different transgenic mouse strains. We typically image the animals at baseline (day 0, after catheter implantation), on day 2 when a mature biofilm is formed, and on day 6 when a mature biofilm is sustained. The here described imaging protocol will be readily translatable for use with other in vivo biofilm models such as the CVC model, for the assessment of antifungal therapies and the validation in transgenic models to study microbial and host factors influencing biofilm formation.
2 Materials
2.1 Candida Albicans Strains
1.
Candida albicans SC5314 (wild-type), a clinical isolate [19]. The genome of this clinical isolate was successfully sequenced.
2.
C. albicans SKCA23 strain (named SKCA23-ACTgLUC) expressing gLUC fused to the endogenous PGA59 gene under the control of ACT1 (actin) promoter (this promoter is active in the yeast as well as hyphal stage of fungal growth). To construct this strain, C. albicans SC5314 was transformed with the Clp10::ACT1p-gLUC59 plasmid [18], which was integrated in the RPS10 locus, in order to constitutively express C. albicans codon-optimized Gaussia princeps luciferase gene (gLUC) at the cell wall. The proper integration was verified by PCR analyses.
2.2 Cell Culture and Media
1.
Standard cell culture medium: RPMI-1640, with l-glutamine and without sodium carbonate buffered with MOPS, pH 7.0.
2.
1× Phosphate buffered saline (for 1 L of 10× PBS: 80 g NaCl, 2 g KCl, 14.4 g Na2HPO4, 2.4 g KH2PO4).
3.
Sterile water.
4.
YPD agar plates (1 % yeast extract, 2 % bacto-peptone, 15 % agar, supplemented with 2 % glucose).
5.
Fetal bovine serum (FBS).
6.
Cell culture plates: 96-well and 24-well black polystyrene plates (Greiner Bio-One, Cellstar®).
7.
Catheters: polyurethane (see Note 1) triple-lumen intravenous catheters (2.4 mm diameter, Certofix Trio S730, BBraun, see Note 2), the triple-lumen part cut into 1 cm long fragments.
8.
Incubator at 37 °C incubator for static incubation of YPD plates.
2.3 Animals
Before you start planning animal experiments, seek approval from your local bioethics committee and follow the respective guidelines for animal handling. Always take care that you carry out all aspects of animal experiments in compliance with national and supranational regulations regarding animal handling and welfare.
1.
Mice: female Balb/C mice 8 weeks of age, kept in individually ventilated filter top cages with free access to standard food and water ad libitum (see Note 3).
2.
Rats: Sprague-Dawley female rats of 200 g (Janvier, France), kept in individually ventilated filter top cages with free access to standard food and water ad libitum.
3.
Immune suppression: suppress the immune system of the animals (see Note 4) 24 h before catheter implant and continue during the entire experiment by adding dexamethasone (0.4 mg/l; Fagron SAS, France) to their drinking water.
4.
During immune suppression, also add antibiotics (e.g., ampicillin sodium powder 0.5 g/l) to the drinking water to avoid possible bacterial infections.
2.4 Anesthesia and Surgery
1.
Anesthesia for mice: prepare 1 ml of anesthetic cocktail by mixing 75 μl of ketamine (Ketamine1000®, 100 mg/ml) with 100 μl of medetomidine (Domitor®, 1 mg/ml) and 825 μl of sterile saline. Administer intraperitoneally (i.p.) 60–80 μl of anesthetic cocktail per 10 g body weight, resulting in a dose of 45–60 mg/kg ketamine and 0.6–0.8 mg/kg medetomidine. For reversal of anesthesia, dilute 50 μl atipamezole (Antisedan®, 5 mg/ml) in 4.95 ml saline, administer i.p. 100 μl per 10 g body weight as antidote, resulting in a dose of 0.5 mg/kg.
2.
Anesthesia for rats: prepare 1 ml of anesthetic cocktail by mixing 600 μl of ketamine with 400 μl of medetomidine. Administer i.p. 100 μl of anesthetic cocktail per 100 g body weight, resulting in a dose of 60 mg/kg ketamine and 0.4 mg/kg medetomidine. For reversal of anesthesia, dilute 1 ml atipamezole in 4 ml saline; administer i.p. 100 μl per 100 g body weight as antidote, resulting in a dose of 1 mg/kg.
3.
Xylocaine gel (2 %) (AstraZeneca) as local anesthetic for the skin.
4.
Terramycin/polymyxin-B ophthalmic ointment (Pfizer), to apply on the eyes to prevent them from drying/infecting while the animal is under anesthesia.
5.
Electric trimmer/razor or disposable razors for shaving the back of the animals.
6.
Clean, sterile surgical tools: fine scissors, tweezers, surgical thread, needle, and tools for suturing the skin.
7.
Sterile gazes, iodo-isopropanol (1 %) as a disinfectant for the skin.
8.
Heating plate or mat to keep the animals warm while they are under anesthesia.
9.
Extra clean cages kept warm (on a warm plate or with a lamp) to keep animals separate until they are fully recovered from anesthesia and surgery.
2.5 BLI
1.
BLI camera: We have used an IVIS 100 system (Caliper LS, Perkin Elmer, USA) and Living Image software (version 2.50.1 for PC, provided by the manufacturer) for measuring and quantifying luminescence in vitro (see Note 5) and in vivo (see Note 6).
2.
Coelenterazine solutions: native coelenterazine (CTZ) was purchased from Nanolight Technologies (USA) and stored at −80 °C according to the manufacturer’s instructions. 5 mg/ml CTZ were dissolved in acidified ethanol according to the manufacturer’s instructions. This stock solution can be stored at −80 °C for the duration of the experiments, typically 6–9 days. For in vitro BLI, 6 μM CTZ working solution was prepared by diluting the stock solution 1:2,000 in sterile PBS. For in vivo BLI, we prepared a 1.2 mM working solution by diluting the stock solution 1:10 in sterile PBS (see Note 7). Foresee 100 μl CTZ working solution per catheter trio. Insulin syringes (0.3 ml) were used for injections. CTZ is light-sensitive, always keep it in the dark (e.g., cover recipients with aluminium foil).
3 Methods
Follow national regulations for handling microbial pathogens class 2. Perform all handlings with open C. albicans suspensions in a vertical laminar air flow cabinet (LAF cabinet) to avoid contamination of cell cultures.
3.1 In Vitro Biofilm Formation
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree