and Soonhag Kim1
(1)
Laboratory of Molecular Imaging, Department of Biomedical Science, College of Life Science, CHA University, Seoul, Republic of Korea
(2)
College of Life Sciences and Biotechnology, Korea University, Seoul, Republic of Korea
Abstract
MicroRNAs (miRNAs) are an abundant class of small noncoding RNA molecules that inhibit the expression of cognate genes in multicellular organisms. These small RNAs have been demonstrated to play crucial roles in a variety of biological processes including cell differentiation, proliferation, and survival. Knowledge of specific expression patterns of miRNAs is critical for functional studies. Here, we describe a bioluminescence reporter gene-based method to measure miRNA activity in cultured cells and mice using a Gaussia luciferase reporter gene controlled by miRNA binding sites in its 3′untranslated region. This method can be used to noninvasively monitor the expression patterns of functionally active miRNAs involved in different biological processes or diseases in mice.
Key words
Bioluminescence reporter gene Gaussia luciferaseMicroRNAImaging1 Introduction
MicroRNAs (miRNAs) are single-stranded, noncoding RNAs of ~22 nucleotides that bind to mRNAs with partial complementarity and induce translational repression and/or mRNA degradation, playing important roles in a wide range of cellular processes [1]. Most miRNA binding sites reside in the 3′ untranslated region (3′UTR) of target mRNAs to mediate posttranscriptional gene regulation [2]. To fully understand the biological functions of miRNAs, it is necessary to determine their spatiotemporal expression patterns. Conventional methods for monitoring the expression of miRNAs include Northern blotting, in situ hybridization, real-time PCR, microarray, and deep sequencing [3]. However, these methods require invasive procedures such as cell disruption and tissue biopsy and thus lack the ability for repetitive detection on the same subjects and in vivo use. More importantly, none of the detection methods do inform whether the expressed miRNA is active for gene silencing. For these reasons, methods to noninvasively visualize both the expression and activity of a given miRNA are desirable.
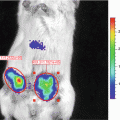
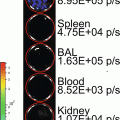
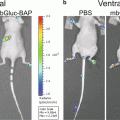
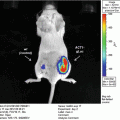
Using an alternative approach first described in Drosophila melanogaster [4], we previously developed a Gaussia luciferase (Gluc) reporter gene-based miRNA imaging system that was designed to detect the expression of functionally active miRNAs in cultured cells and mice. Gluc is much more sensitive than commonly used luciferases from firefly or Renilla and catalyzes the oxidation of the substrate coelenterazine in a reaction that emits light with a peak wavelength of 480 nm [5]. The reporter gene has the Gluc coding sequence fused to a 3′UTR bearing perfectly complementary binding sites for the miRNA, and thus its expression is controlled by miRNA-directed cleavage of the Gluc mRNA through the RNA interference (RNAi) pathway [6, 7] (Fig. 1). Accordingly, an increase in the activity of the target miRNA is reflected by a decrease in Gluc activity. The Gluc reporter is constitutively expressed under control of the cytomegalovirus (CMV) promoter in cells lacking the target miRNA. In contrast, where present and active, an miRNA represses the expression of the reporter containing its perfectly complementary binding sites by RNAi in a quantitative manner. We have successfully used this approach to detect the activity of distinct miRNAs in vitro and in vivo whose expression is modulated during differentiation or in tumor cells [8–12]. For example, up-regulation of miR-9, miR-23a, and miR-124a during neuronal differentiation of a mouse embryonic carcinoma cell line (P19 cells) resulted in a decrease in the bioluminescent signal derived from the corresponding miRNA-specific Gluc reporters, indicating that the Gluc activity depends on the levels of functionally active miRNAs [8, 9, 11].
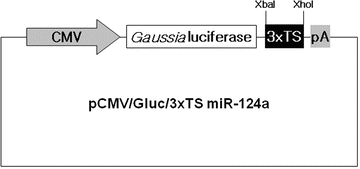

Fig. 1
The schematic diagram of a pCMV/Gluc/3×TS miR-124a construct. The Gluc reporter gene contains three copies of a perfect target site for mature miR-124a (3×TS miR-124a) in the 3’UTR. The expression of the reporter is repressed as miR-124a is targeted to 3×TS of the Gluc mRNA
In this chapter, we exemplify bioluminescence reporter gene-based detection of miR-124a activity in culture and in vivo using P19 cells in which miR-124a is gradually up-regulated during neuronal differentiation [11, 13]. We describe detailed protocols for construction of a miR-124a-specific Gluc reporter plasmid and for bioluminescence imaging of miR-124a activity in cell lysates and in nude mice implanted with Gluc-expressing cells.
2 Materials
Prepare all solutions using ultrapure water (18 MΩ dH2O) and analytical grade reagents. Store all reagents at −20 °C, unless otherwise specified. Follow institutional regulations for waste disposal and animal experiments.
2.1 Recombinant DNA Technique Components
1.
Gaussia luciferase-expressing vector, pCMV GLUC-1 (Targeting Systems, El Cajon, CA, USA) (see Note 1).
2.
Firefly luciferase-expressing vector (pCMV/Fluc), pGL3-Basic (Promega, Madison, WI, USA) (see Note 2).
3.
3xTS miR-124a (three tandem copies of a perfect target site for mature miR-124a, which are separated by a spacer of five nucleotides in between) sense and antisense oligonucleotides that once annealed have XbaI- and XhoI-compatible ends (see Note 3): 100 μM sense and antisense 3xTS miR-124a oligonucleotides each dissolved in nuclease-free water. 3xTS miR-124a sense (5′-TCGAGaatctagtTGGCATTCACCGCGTGCCTTAA tagta TGGCATTCACCGCGTGCCTTAA tagta TGGCATTCACCGCGTGCCTTAA T-3′; bold, miR-124a target site; underlined, XbaI 5′-protruding end and XhoI 3′-recessive end; italic lowercase, spacer sequence). 3xTS miR-124a antisense (5′-CTAGA TTAAGGCACGCGGTGAATGCCA tacta TTAAGGCACGCGGTGAATGCCA tacta TTAAGGCACGCGGTGAATGCCAactagattC-3′; bold, miR-124a target site; underlined, XhoI 5′-protruding end and XbaI 3′-recessive end; italic lowercase, spacer sequence).
4.
Annealing buffer (10×): 100 mM Tris–HCl, pH 7.5, 1 M NaCl, and 10 mM EDTA.
5.
Restriction enzymes XbaI and XhoI.
6.
Alkaline phosphatase, shrimp.
7.
T4 DNA ligase.
9.
LB broth: 0.01 % Bacto-tryptone, 0.005 % Bacto-yeast extract, and 0.01 % NaCl (see Note 5). Store at 4 °C.
10.
LB agar plates containing ampicillin: 0.01 % Bacto-tryptone, 0.005 % Bacto-yeast extract, 0.01 % NaCl, 1.5 % Bacto-agar, and 100 μg/mL ampicillin (see Note 5). Store at 4 °C.
11.
Electrophoresis-grade agarose. Store at room temperature.
12.
Agarose gel electrophoresis equipment.
14.
Gel-loading buffer (10×): 0.25 % bromophenol blue, 0.25 % xylene cyanol FF, and 50 % glycerol. Store at 4 °C.
15.
DNA molecular weight markers.
16.
TAE buffer for agarose gel electrophoresis (50×): Store at room temperature.
17.
UV transilluminator.
18.
T7 primer: 5′-TAATACGACTCACTATAGGG-3′.
19.
CMV/reverse 2 primer: 5′-CTTACTTGGCATGACAGTAA-3′.
20.
Taq DNA polymerase.
21.
dNTPs: A mixture of dATP, dCTP, dGTP, and dTTP, each at 100 nM.
22.
PCR machine.
23.
Incubator and orbital shaker for bacterial culture.
24.
NucleoBond Xtra Midi kit (Macherey-Nagel, Düren, Germany). Store at room temperature.
2.2 Culture and Transfection of P19 Cells, and In Vitro Luciferase Assay Components
1.
P19 cells (a mouse embryonic carcinoma cell line, ATCC no. CRL-1825).
2.
150 mm tissue culture dishes and 6-well plates.
3.
Gelatin.
4.
A laminar flow cell culture hood and 37 °C incubator with humidified atmosphere of 5 % CO2.
5.
Trypan blue: 0.5 % in phosphate-buffered saline (PBS).
6.
Hemocytometer.
7.
Inverted microscope.
8.
Growth medium: Minimal essential medium alpha (α-MEM) supplemented with 7.5 % bovine calf serum, 2.5 % fetal bovine serum, and 1 % Antibiotic–Antimycotic solution. Store at 4 °C, which is stable for <3 months.
9.
Lipofectamine and Plus reagent (Invitrogen). Store at 4 °C.
10.
Opti-MEM medium (GIBCO, Grand Island, NY, USA). Store at 4 °C.
11.
Dulbecco’s phosphate-buffered saline. Store at 4 °C.
12.
Neuronal differentiation medium: Dulbecco’s Modified Eagle’s medium (DMEM)/F12 [1:1] supplemented with 1 % insulin–transferrin–selenium, 1 % Antibiotic–Antimycotic solution, and 0.5 μM all-trans-retinoic acid (RA) (see Note 7). Store at 4 °C, which is stable for <3 months.