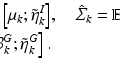Fig. 1.
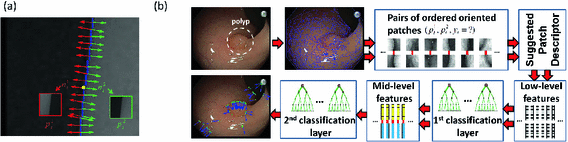
Significant variation in visual characteristics of polyps. (a) Color and appearance variation of the same polyp due to varying lighting conditions. (b) Texture and shape variation among polyps. Note how the distance between the polyps and colonoscopy camera determines the availability of polyp texture. (c) Other polyp-like structures in the colonoscopic view (Color figure online).
However, designing a high-performance system for computer-aided polyp detection is challenging: (1) Polyps appear differently in color, and even the same polyp, as shown in Fig. 1(a), may look differently due to varying lighting conditions. (2) Polyps have large inter- and intra-morphological variations. As shown in Fig. 1(b), the shapes of polyps vary considerably from one to another. The intra-shape variation of polyps is caused by various factors, including the viewing angle of the camera and the spontaneous spasms of the colon. (3) Visibility of the texture on the surface of polyps is also varying due to biological factors and distance between the polyps and the colonoscopy camera. This can be seen in Fig. 1(b) where texture visibility decrease as the polyps distance from the capturing camera. The significant variations among polyps suggest that there is no single feature that performs the best for detecting all the polyps.
As a result, to achieve a reliable polyp detection system, it is critical to fuse all possible features of polyps, including shapes, color, and texture. Each of these features has strengths and weaknesses. Among these features, geometric shapes are most robust because polyps, irrespective of their morphology and varying levels of protrusion, have at least one curvilinear head at their boundaries. However, this property is not highly specific to polyps. This is shown in Fig. 1(c) where non-polyp structures exhibit similar geometric characteristics to polyps. Texture features have the weakness of limited availability; however, when visible, they can distinguish polyps from some non-polyp structures such as specular spots, dirt, and fecal matter. In addition, temporal information is available in colonoscopy and may be utilized to distinguish polyps from bubbles or other artifacts that only briefly appear in colonoscopy videos.
Our key contribution of this paper is an idea to exploit the strengths of each feature and minimize its weaknesses. To realize this idea, we have developed a new system for polyp detection with two stages. The first stage builds on the robustness of shape features of polyps to reliably generate a set of candidate detections with a high sensitivity, while the second stage utilizes the high discriminative power of the computationally expensive features to effectively reduce false positive detections. More specifically, we employ a unique edge classifier coupled with a voting scheme to capture geometric features of polyps in context and then harness the power of convolutional deep networks in a novel score fusion approach to capture shape, color, texture, and temporal information of the candidates. Our experimental results based on the largest annotated polyp database demonstrate that our system achieves high sensitivity to polyps and generates significantly less number of false positives compared to state-of-the-art. This performance improvement is attributed to our reliable candidate generation and effective false positive reduction methods.
2 Related Works
Automatic polyp detection in colonoscopy videos has been the subject of research for over a decade. Early methods, e.g., [1, 3] for detecting colonic polyps utilized hand-crafted texture and color descriptors such as LBP and wavelet transform. However, given large color variation among polyps and limited texture availability on the surface of polyps (See Fig. 1), such methods could offer only a partial solution. To avoid such limitations, more recent techniques have considered temporal information [6] and shape features [2, 7, 9–11], reporting superior performance over the early polyp detection systems. Despite significant advancements, state-of-the-art polyp detection methods fail to achieve a clinically acceptable performance. For instance, to achieve the polyp sensitivity of 50 %, the system suggested by Wang et al. [11] generates 0.15 false positives per frame or approximately 4 false positive per second. Similarly, the system proposed in [10], which is evaluated on a significantly larger dataset, generates 0.10 false positives per frame. Clearly, such systems that rely on a subset of polyp characteristics are not clinically viable—a limitation that this paper aims to overcome.
3 Proposed Method
Our computer-aided polyp detection system is designed based on our algorithms [7, 8, 10], consisting of 2 stages where the first stage utilizes geometric features to reliably generate polyp candidates and the second stage employs a comprehensive set of deep features to effectively remove false positives. Figure 2 shows a schematic overview of the suggested method.
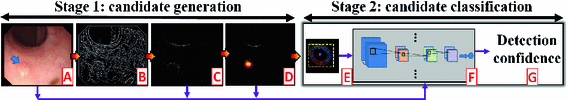

Fig. 2.
Our system consists of 2 stages: candidate generation and classification. Given a colonoscopy frame (A), we first obtain a crude set of edge pixels (B). We then refine this edge map using a classification scheme where the goal is to remove as many non-polyp boundary pixels as possible (C). The geometric features of the retained edges are then captured through a voting scheme, generating a voting map whose maximum indicates the location of a polyp candidate (D). In the second stage, a bounding box is estimated for each generated candidate (E) and then a set of convolution neural networks—each specialized in one type of features—are applied in the vicinity of the candidate (F). Finally, the CNNs are aggregated to generate a confidence value (G) for the given polyp candidate.
3.1 Stage 1: Candidate Generation
Our unique polyp candidate generation method exploits the following two properties: (1) polyps have distinct appearance across their boundaries, (2) polyps, irrespective of their morphology and varying levels of protrusion, feature at least one curvilinear head at their boundaries. We capture the first property with our image characterization and edge classification schemes, and capture the second property with our voting scheme.
Constructing Edge Maps. Given a colonoscopy image, we use Canny’s method to extract edges from each input channel. The extracted edges are then put together in one edge map. Next, for each edge in the constructed edge map, we determine edge orientation. The estimated orientations are later used for extracting oriented patches around the edge pixels.
Image Characterization. Our patch descriptor begins with extracting an oriented patch around each edge pixel. The patch is extracted so that the containing boundary is placed vertically in the middle of the patch. This representation allows us to capture desired information across the edges independent of their orientations. Our method then proceeds with forming  sub-patches all over the extracted patch. Each sub-patch has 50 % overlap with the neighboring sub-patches. For a compact representation, we compress each sub-patch into a 1D signal S by averaging intensity values along each column. We then apply a 1D discrete cosine transform (DCT) to the resulting signal:
sub-patches all over the extracted patch. Each sub-patch has 50 % overlap with the neighboring sub-patches. For a compact representation, we compress each sub-patch into a 1D signal S by averaging intensity values along each column. We then apply a 1D discrete cosine transform (DCT) to the resulting signal:
![$$\begin{aligned} C_k=\frac{2}{n}w(k)\sum _{i=0}^{n-1}S[i]\cos (\frac{2i+1}{2n}\pi k) \end{aligned}$$](/wp-content/uploads/2016/09/A339424_1_En_25_Chapter_Equ1.gif)
where
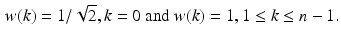 With the DCT, the essential information of the intensity signal can be summarized in a few coefficients. We discard the DC component
With the DCT, the essential information of the intensity signal can be summarized in a few coefficients. We discard the DC component 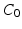 because the average patch intensity is not a robust feature—it is affected by a constant change in patch intensities. However, the next 3 DCT coefficients
because the average patch intensity is not a robust feature—it is affected by a constant change in patch intensities. However, the next 3 DCT coefficients 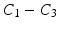 are more reliable and provide interesting insight about the intensity signal.
are more reliable and provide interesting insight about the intensity signal. 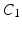 measures whether the average patch intensity along the horizontal axis is monotonically decreasing (increasing) or not,
measures whether the average patch intensity along the horizontal axis is monotonically decreasing (increasing) or not, 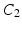 measures the similarity of the intensity signal against a valley (ridge), and finally
measures the similarity of the intensity signal against a valley (ridge), and finally 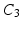 checks for the existence of both a valley and a ridge in the signal. The higher order coefficients
checks for the existence of both a valley and a ridge in the signal. The higher order coefficients 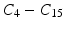 may not be reliable for feature extraction because of their susceptibility to noise and other degradation factors in the images.
may not be reliable for feature extraction because of their susceptibility to noise and other degradation factors in the images.
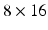 sub-patches all over the extracted patch. Each sub-patch has 50 % overlap with the neighboring sub-patches. For a compact representation, we compress each sub-patch into a 1D signal S by averaging intensity values along each column. We then apply a 1D discrete cosine transform (DCT) to the resulting signal:
sub-patches all over the extracted patch. Each sub-patch has 50 % overlap with the neighboring sub-patches. For a compact representation, we compress each sub-patch into a 1D signal S by averaging intensity values along each column. We then apply a 1D discrete cosine transform (DCT) to the resulting signal:![$$\begin{aligned} C_k=\frac{2}{n}w(k)\sum _{i=0}^{n-1}S[i]\cos (\frac{2i+1}{2n}\pi k) \end{aligned}$$](/wp-content/uploads/2016/09/A339424_1_En_25_Chapter_Equ1.gif)
(1)
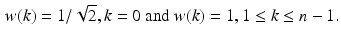
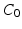 because the average patch intensity is not a robust feature—it is affected by a constant change in patch intensities. However, the next 3 DCT coefficients
because the average patch intensity is not a robust feature—it is affected by a constant change in patch intensities. However, the next 3 DCT coefficients 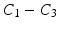 are more reliable and provide interesting insight about the intensity signal.
are more reliable and provide interesting insight about the intensity signal. 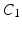 measures whether the average patch intensity along the horizontal axis is monotonically decreasing (increasing) or not,
measures whether the average patch intensity along the horizontal axis is monotonically decreasing (increasing) or not, 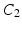 measures the similarity of the intensity signal against a valley (ridge), and finally
measures the similarity of the intensity signal against a valley (ridge), and finally 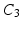 checks for the existence of both a valley and a ridge in the signal. The higher order coefficients
checks for the existence of both a valley and a ridge in the signal. The higher order coefficients 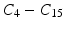 may not be reliable for feature extraction because of their susceptibility to noise and other degradation factors in the images.
may not be reliable for feature extraction because of their susceptibility to noise and other degradation factors in the images.The selected DCT coefficients 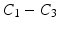 are still undesirably proportional to linear illumination scaling. We therefore apply a normalization treatment. Mathematically,
are still undesirably proportional to linear illumination scaling. We therefore apply a normalization treatment. Mathematically,
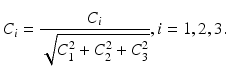 Note that we use the norm of the selected coefficients for normalization rather than the norm of entire DCT coefficients. By doing so, we can avoid the expensive computation of all the DCT components. The final descriptor for a given patch is obtained by concatenating the normalized coefficients selected from each sub-patch.
Note that we use the norm of the selected coefficients for normalization rather than the norm of entire DCT coefficients. By doing so, we can avoid the expensive computation of all the DCT components. The final descriptor for a given patch is obtained by concatenating the normalized coefficients selected from each sub-patch.
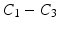 are still undesirably proportional to linear illumination scaling. We therefore apply a normalization treatment. Mathematically,
are still undesirably proportional to linear illumination scaling. We therefore apply a normalization treatment. Mathematically,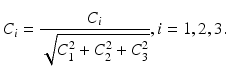
The suggested patch descriptor has 4 advantages. First, our descriptor is fast because compressing each sub-patch into a 1D signal eliminates the need for expensive 2D DCT and that only a few DCT coefficients are computed from each intensity signal. Second, due to the normalization treatment applied to the DCT coefficients, our descriptor achieves invariance to linear illumination changes, which is essential to deal with varying lighting conditions (see Fig. 1). Third, our descriptor is rotation invariant because the patches are extracted along the dominant orientation of the containing boundary. Fourth, our descriptor handles small positional changes by selecting and averaging overlapping sub-patches in both horizontal and vertical directions.
Edge Classification. Our classification scheme has 2 layers. In the first layer, we learn a discriminative model to distinguish between the boundaries of the structures of interest and the boundaries of other structures in colonoscopy images. The structures of interest consists of polyps, vessels, lumen areas, and specular reflections. Specifically, we collect a stratified set of 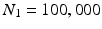 oriented patches around the boundaries of structures of interest and r andom structures in the training images,
oriented patches around the boundaries of structures of interest and r andom structures in the training images, 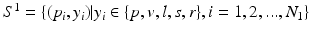 . Once patches are extracted, we train a five-class random forest classifier with 100 fully grown trees. The resulting probabilistic outputs can be viewed as the similarities between the input patches and the predefined structures of interest. Basically, the first layer receives low-level image features from our patch descriptor and then produces mid-level semantic features.
. Once patches are extracted, we train a five-class random forest classifier with 100 fully grown trees. The resulting probabilistic outputs can be viewed as the similarities between the input patches and the predefined structures of interest. Basically, the first layer receives low-level image features from our patch descriptor and then produces mid-level semantic features.
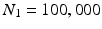 oriented patches around the boundaries of structures of interest and r andom structures in the training images,
oriented patches around the boundaries of structures of interest and r andom structures in the training images, 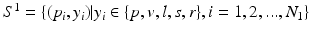 . Once patches are extracted, we train a five-class random forest classifier with 100 fully grown trees. The resulting probabilistic outputs can be viewed as the similarities between the input patches and the predefined structures of interest. Basically, the first layer receives low-level image features from our patch descriptor and then produces mid-level semantic features.
. Once patches are extracted, we train a five-class random forest classifier with 100 fully grown trees. The resulting probabilistic outputs can be viewed as the similarities between the input patches and the predefined structures of interest. Basically, the first layer receives low-level image features from our patch descriptor and then produces mid-level semantic features.In the second layer, we train a 3-class random forest classifier with 100 fully grown trees. Specifically, we collect 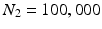 pairs of oriented patches, of which half are randomly selected from the polyp boundaries and the rest are selected from random non-polyp edge segments. For an edge pixel at angle
pairs of oriented patches, of which half are randomly selected from the polyp boundaries and the rest are selected from random non-polyp edge segments. For an edge pixel at angle 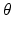 , one can obtain two oriented image patches
, one can obtain two oriented image patches 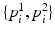 by interpolating the image along the two possible normal directions
by interpolating the image along the two possible normal directions 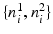 . As shown in Fig. 3(a), for an edge pixel on the boundary of a polyp, only one of the normal directions points to the polyp region. Our classification scheme operates on each pair of patches with two objectives: (1) to classify the underlying edge into polyp and non-polyp categories, and (2) to determine the desired normal direction among
. As shown in Fig. 3(a), for an edge pixel on the boundary of a polyp, only one of the normal directions points to the polyp region. Our classification scheme operates on each pair of patches with two objectives: (1) to classify the underlying edge into polyp and non-polyp categories, and (2) to determine the desired normal direction among 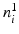 and
and 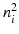 such that it points towards the polyp location. Henceforth, we refer to the desired normal direction as “voting direction”.
such that it points towards the polyp location. Henceforth, we refer to the desired normal direction as “voting direction”.
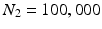 pairs of oriented patches, of which half are randomly selected from the polyp boundaries and the rest are selected from random non-polyp edge segments. For an edge pixel at angle
pairs of oriented patches, of which half are randomly selected from the polyp boundaries and the rest are selected from random non-polyp edge segments. For an edge pixel at angle 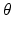 , one can obtain two oriented image patches
, one can obtain two oriented image patches 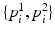 by interpolating the image along the two possible normal directions
by interpolating the image along the two possible normal directions 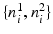 . As shown in Fig. 3(a), for an edge pixel on the boundary of a polyp, only one of the normal directions points to the polyp region. Our classification scheme operates on each pair of patches with two objectives: (1) to classify the underlying edge into polyp and non-polyp categories, and (2) to determine the desired normal direction among
. As shown in Fig. 3(a), for an edge pixel on the boundary of a polyp, only one of the normal directions points to the polyp region. Our classification scheme operates on each pair of patches with two objectives: (1) to classify the underlying edge into polyp and non-polyp categories, and (2) to determine the desired normal direction among 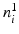 and
and 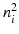 such that it points towards the polyp location. Henceforth, we refer to the desired normal direction as “voting direction”.
such that it points towards the polyp location. Henceforth, we refer to the desired normal direction as “voting direction”.
Fig. 3.




(a) A pair of image patches 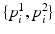 extracted from an edge pixel. The green and red arrows show the two possible normal directions
extracted from an edge pixel. The green and red arrows show the two possible normal directions 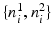 for a number of selected edges on the displayed boundary. The normal directions are used for patch alignment. (b) The suggested edge classification scheme given a test image. The edges that have passed the classification stage are shown in green. The inferred normal directions are visualized with the blue arrows for a subset of the retained edges (Color figure online).
for a number of selected edges on the displayed boundary. The normal directions are used for patch alignment. (b) The suggested edge classification scheme given a test image. The edges that have passed the classification stage are shown in green. The inferred normal directions are visualized with the blue arrows for a subset of the retained edges (Color figure online).
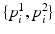 extracted from an edge pixel. The green and red arrows show the two possible normal directions
extracted from an edge pixel. The green and red arrows show the two possible normal directions 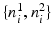 for a number of selected edges on the displayed boundary. The normal directions are used for patch alignment. (b) The suggested edge classification scheme given a test image. The edges that have passed the classification stage are shown in green. The inferred normal directions are visualized with the blue arrows for a subset of the retained edges (Color figure online).
for a number of selected edges on the displayed boundary. The normal directions are used for patch alignment. (b) The suggested edge classification scheme given a test image. The edges that have passed the classification stage are shown in green. The inferred normal directions are visualized with the blue arrows for a subset of the retained edges (Color figure online).Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree