© Springer Science+Business Media New York 2015
Lin He (ed.)Mass Spectrometry Imaging of Small MoleculesMethods in Molecular BiologyMethods and Protocols120310.1007/978-1-4939-1357-2_11. Current Status and Future Prospects of Mass Spectrometry Imaging of Small Molecules
(1)
Department of Chemistry, North Carolina State University, 2620 Yarbrough Drive, CB 8204, Raleigh, NC 27695, USA
Abstract
In the field of small-molecule studies, vast efforts have been put forth in order to comprehensively characterize and quantify metabolites formed from complex mechanistic pathways within biochemical and biological organisms. Many technologies and methodologies have been developed to aid understanding of the inherent complexities within biological metabolomes. Specifically, mass spectroscopy imaging (MSI) has emerged as a foundational technique in gaining insight into the molecular entities within cells, tissues, and whole-body samples. In this chapter we provide a brief overview of major technical components involved in MSI, including topics such as sample preparation, analyte ionization, ion detection, and data analysis. Emerging applications are briefly summarized as well, but details will be presented in the following chapters.
Key words
Mass spectroscopy imaging (MSI)MetaboliteSample preparationMS ionizationMS analyzerMetabolites are the class of low-molecular-weight molecules resultant from metabolic processes within biological systems. The quantitative and qualitative studies of metabolites are designed to enhance our existing knowledge of the metabolome, the intricate compilation of metabolites within cells, tissues, biofluids, and organs found in eukaryotic organisms. Characterization of global response patterns, metabolic pathways, and cell-specific functions in plant and animal metabolites provides insight into cellular and physiological changes in response to genetic or environmental stressors and is of great importance in the fields of agriculture, pharmacology, and molecular medicine [1–4].
Ever since the bloom of metabolomics studies, mass spectrometry (MS) has been the method of choice for the field, owing to its unprecedented resolving power and sensitivity [5, 6]. Providing snapshots of spatial distribution of biologically relevant metabolites in complex samples, mass spectrometry imaging (MSI) provides additional dimension of information to metabolite profiling hence it enables direct 2D visualization, and subsequent correlation, of their concerted regulatory roles in metabolic network [7–10]. Strategic fingerprinting each chemical component based on its unique molecular composition, MSI has become the workhorse technique in gathering heterogenic topological information from biological samples to elucidate metabolite pathways associated with various biochemical processes that are multifarious in nature.
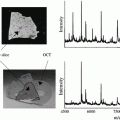
In a typical MSI experiment setting, samples are canvassed in a raster of points, each of which has a corresponding x and y coordinates (sometimes with the axis z monitored as well). As the sampling beam scans over the sample, an independent mass spectrum is collected for the predefined mass range. Spectra for each raster point, or x–y coordinate, are then processed and reconstructed into a 2D (or 3D) molecular image of the system being analyzed. Achieving universal detection and quantification of small molecules simultaneously, however, is a nontrivial task. A robust workflow with carefully designed sample preparation, data management, and quantification strategies is a must. First and foremost, regardless of the sample types, properly handling and preparation are critical to ensure preservation of the molecular morphology and chemical integrity for reproducible and reliable results [11–13]. For example, physical (low temperature) or chemical (fixation reagents) means have been used to suppress metabolite degradation. Cleverly designed and carefully executed strategies have been developed to process natural samples of great variations (e.g., patient to patient, diet, age, disease state), which are reiterated with great emphasis throughout this book.
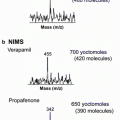
Nonbiased liberation of analytes from a solid surface into gas phase, with well-preserved spatial locations and relative concentrations, for subsequent MS analysis is the next critical step in MSI experiments. Breakthroughs and improvements in various desorption and ionization techniques have significantly broadened the repertoire of ionization sources for MSI. Common ionization methods used in MSI include secondary ion mass spectrometry (SIMS), matrix-assisted laser/desorption ionization (MALDI), desorption electrospray ionization (DESI), and their derivatives, all providing a multitude of advantageous features in their own way [7, 8].
Ionization through SIMS entails a high-energy primary ion beam to generate secondary ions from a surface [14–16]. It has long been used for elemental analysis of thin films and materials because the ion beam can be easily focused in an electric field with submicron resolution. Successful extension to small-molecule profiling in cellular and tissue imaging has been demonstrated along with the new generation of ion beams (e.g., C60 + or Au cluster ion beam) and has become one of the routine approaches in subcellular 2-D and 3-D imaging [16].
The acclaim-to-fame of MALDI is its effective ionization of large molecules, along with electrospray ionization, that changes the landscape of proteomics studies. Its applications to small-molecule analysis was hindered initially by the presence of high matrix background but it becomes less of an issue in recent years with rapid advancement of high-resolution mass analyzers [17–19]. For MALDI and other ionization sources that involve matrix-coated samples, choosing an appropriate matrix is vital for a successful measurement [20, 21]. Subsequent matrix deposition is another important step to ensure reproducible matrix coating and uniform crystal formation, which has direct effects on limiting analyte migration and preserving spatial resolution and ion intensities [20–23]. Techniques, both manual and automated, have been created to achieve optimum reproducibility of matrix deposition for MSI experiments. Successful examples include dried-droplet, pneumatic nebulization, sublimation, spray-coating, acoustic deposition, ChIP, electrospray, solvent-free, and chemical inkjet printing [24–29]. While these approaches share the same goal, each differs in the size of matrix crystals formed, economic feasibility, practicality, and throughput, so a thorough inspection of resulting samples is necessary in order to choose the most appropriate method for a given MSI experiment. Despite the arduous nature of confining each component in matrix deposition, utilizing MALDI-MSI has explicit advantages for metabolite imaging: its versatility has been shown in the diverse subjects studied, such as whole-body samples, cancerous tissue diagnostics, and drug development [6, 7, 30, 31].
Development of ambient-based MSI techniques offers a more effective solution to the issues associated with sample stability under vacuum. In this regard, DESI has established itself as a versatile and dynamic ambient ionization source with considerable capabilities for metabolite imaging [13, 32]. As one of the most widely used ionization methods nowadays, DESI has minimized potential matrix interference in small-molecule detection [32–34] and has been successfully utilized in imaging whole-body sections, profiling compounds in forensic studies, detecting antifungal metabolites within natural products, monitoring metabolic species in plant imprints, and characterizing varying disease states within tissue samples. While the large scanning footprint due to the size of the incoming charged droplet beam is one of the trade-off of DESI, various improvements to reduce beam size and better ionization selectivity, as discussed in later chapters, have made great strides in high-resolution MS imaging [13, 35, 36].
Taking advantages of nanotechnology development, novel nanomaterials have been incorporated in various means for analyte ionization with better ionization efficiency and minimized sample preparation [37–40]. Desorption/ionization on silicon (DIOS) was the most successful one based on porous silicon substrate. The more effective version of DIOS by facilitating the direct and unbiased detection and imaging of metabolites within biofluids and biological tissues [41, 42]. Substrates such as porous metal oxides, metal nanoparticles, carbon nanotubes, and thin films have all been implemented as viable surface pretreatment options, either individually or in conjunction with a matrix-based application, to optimize results in MSI experiments.
Needless to say, although a plethora of ionization methods have been developed and are presented in this protocol book, selection of the appropriate method for specific samples is decided by desired imaging results, as each ionization source possesses its own intrinsic benefits and limitations in the achievable spatial resolution, mass range and sensitivity, etc. This book includes a handful of examples on how different ionization methods are used in analyzing similar samples and vice versa, and more importantly, the considerations took place behind such decisions.
Upon successful releasing of analytes into gas phase, an additional element is required for any sound and successful metabolite imaging experiment—an efficient mass analyzer [6, 43]. In the broadest sense, mass analyzers function as a high precision balance by differentiating molecules and molecular fragments based on their mass-to-charge (m/z) ratios. The most widely used ones are time-of-flight (TOF), Fourier transform ion cyclotron resonance (FTICR), and Orbitrap mass analyzers. Time-of-flight (TOF) mass analyzers detect ions based on their kinetic energy in either a linear or reflectron mode. The latter mode uses electrostatic mirrors to adjust for slight differences in kinetic energy after the desorption/ionization event, which leads to a pronounced improvement in mass resolving power [43]. Tandem MS analyzers in the form of TOF–TOF provide additional capability for pertinent structural elucidation of analytes through extensive fragmentation. Together with its high throughput, superior sensitivity, and large m/z range, TOFs are the predominant mass analyzers used in MSI experiments.
Other mass analyzers such as Fourier transform ion cyclotron resonance (FTICR) and ion-trap/orbitrap have become more and more popular for their high mass accuracy, imperative features for ambiguous distinction of minuscule mass differences [6, 43]. FTICR is much different from TOF mass analyzers as detection of analytes is based on the frequencies of the ions traveling in orbits. FTICR is advantageous for metabolite imaging purposes as it offers high mass measurement accuracy (MMA), high resolving power, and multiplexed MS/MS capabilities, but at the price of lower throughput when compared to TOF [44, 45]. The applications, but due to their low mass accuracy they are frequently coupled to another mass analyzer to enhance performance [46, 47]. Orbitrap is another modern MS analyzer that provides excellent mass resolving power and mass accuracy, comparable to FTICR. In a simplistic sense, orbitraps function by confining ions around a central electrode. The axial oscillations of these ions are translated into an image current, where the m/z ratios of the ions can then be determined. Orbitraps are most often coupled to linear ion traps (LITs) mass analyzers for general metabolomic studies orbitraps to achieve fast scan speeds and sensitive detection, despite low ion abundance levels [46– 49].
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree