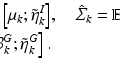accuracy by hashing and retrieving multiple cues of half-million cells.
1 Introduction
Content-based image retrieval (CBIR) has been an effective approach in analyzing medical images [1–3]. It aims to retrieve and visualize relevant medical images with diagnosis information, which assist doctors to make consistent clinical decisions. Successful use cases include clinical pathology, mammogram analysis, and categorization of X-ray images [1, 4–7]. Recently, the research focus of CBIR for medical images has been on the efficient and large-scale methods, and several benchmarks have been designed, such as Image CLEF and VISCERAL [2, 8]. The motivation of leveraging large databases of training images is that they have the potential to offer abundant information to precisely interpret the new data. However, the scalability or efficiency of these algorithms is usually an issue. In this paper, we design a scalable CBIR algorithm to tackle a challenging problem, differentiating lung cancers using histopathological images. Lung cancer is one of the most common cancers [9]. There are four typical histologic types of lung cancers, including adenocarcinoma, squamous carcinoma, small cell carcinoma, and large cell carcinoma, each of which needs a different treatment [10]. Therefore, early diagnosis and differentiation of these four types is clinically important. Bronchial biopsy is one of the most effective diagnosis methods to differentiate them, with the aid of Computer Aided Diagnosis (CAD) systems [11–13]. Many previous methods emphasize the diagnosis of small cell vs. non-small cell (i.e., adenocarcinoma, squamous carcinoma, and large cell carcinoma) types of lung cancers, achieving promising accuracy. On the other hand, differentiation of the adenocarcinoma and squamous carcinoma, both of which are non-small cells, is much more difficult, while it is also clinically significant, since management protocols of these two types of cancers are different [14]. This is challenging because the difference between the adenocarcinoma and squamous carcinoma highly depends on the cell-level information, e.g., its morphology, texture and appearance, requiring to analyze multiple cues of all cells for accurate diagnosis. In fact, thoroughly examining cell-level information is necessary for many use cases of biopsy or histopathological image analysis. To this end, a potential solution proposed recently is to extract high-dimensional local texture features (e.g., SIFT [15]) that align with the cell-level information, and then compress them as binary codes via hashing-based CBIR algorithms [16] to improve the computational efficiency. Hashing methods [17–20] have been intensively investigated in machine learning communities, which enable fast approximated nearest neighbors (ANN) search for scalability. However, information loss is inevitable in such holistic approximation of cell-level information. Segmenting and hashing each cell offers a potential solution [21], and our framework also follows this strategy. Nonetheless, it only utilizes one type of features, while multiple cues should be examined for accurate classification.
Different from these previous methods, our proposed framework is able to (1) hash multiple cues of all cells with weights, and (2) accommodate new training data on-the-fly. Specifically, each cue of a cell is represented as binary codes through supervised hashing with kernels [20]. Then, the importance of such hash entry is determined adaptively according to its discriminativity for differentiating different classes, based on several measures such as probability scores. Given these probability scores, we integrate multiple features by considering importance. An additional benefit of this design is that the hashing results of multiple cues can be updated on-the-fly when handling new training samples. The classification of the whole image is conducted by aggregating the results of multiple cues of all cells. We evaluate our algorithm on this specific problem of differentiating lung cancers, using a large dataset containing 1120 lung microscopic tissue images acquired from hundreds of patients, achieving an accuracy of 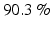 within several seconds by searching among half-million cells.
within several seconds by searching among half-million cells.
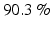 within several seconds by searching among half-million cells.
within several seconds by searching among half-million cells.The rest of the paper is organized as follows. Section 2 presents our framework for real-time cell examination by hashing multiple cues of cells, including details of the hashing method, probabilistic-based weighting schemes, and aggregation of multiple cues. Section 3 shows the experimental results and comparisons. Section 4 draws the concluding remarks and shows future directions.
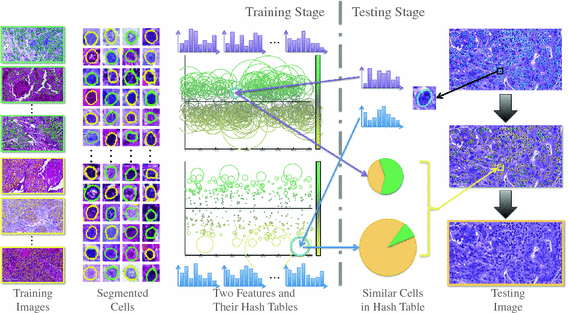

Fig. 1.
Overview of our framework. In the training stage, the input is histopathological images representing two types of lung cancers. Green stands for adenocarcinoma, and yellow stands for squamous carcinoma. First, all cells are detected and segmented from these images. Second, two types of texture features are extracted and compressed as binary codes by hashing methods. These hash codes are visualized in two hash tables, representing two features, according to their ability to differentiate two categories (details in Sect. 2.3). In the testing stage, all cells are segmented from the query image, from which feature and binary codes are obtained using the same preprocess. Each cell is mapped into hash tables to search for the most similar cases, which are used to interpret the category of this unknown cell. The hash entries of two features are then integrated to enhance the accuracy. Finally, the results of all cells are aggregated to classify the testing image (Color figure online).
2 Methodology
2.1 Overview
The overview of our proposed framework is illustrated in Fig. 1. It includes automatic cell detection and segmentation, supervised hashing that generates binary codes from multiple types of image features, and the probabilistic-based weighting scheme that decides the importance of the hash entry for each cell. Specifically, the segmentation is based on the off-the-shelf method [22], while many other methods and systems are also applicable for this task [23, 24]. After segmenting all cells from the training images, a large-scale database of half-million cell patches is created. Then, two types of texture features [25, 26] are extracted for each cell, within the segmented region. After that, kernelized and supervised hashing (KSH) [20] is employed as a baseline to compress these features as binary code, since it can bridge the semantic gap of image appearances and their labels, which is essential for medical image retrieval. However, different from hashing the whole image, hashing cells (i.e., sub-regions of the whole image) is more challenging, due to cells’ high intra-class but low inter-class variations. Therefore, traditional hashing methods result in low-discriminative hash entries. In addition, it is necessary to integrate multiple features from each cell, so the information can be largely explored. Our solution is the probabilistic-based weighting schemes that stress discriminative hash entries, and the integration of multiple cues of cells through the probability scores. Given a testing image, the same framework is utilized to segment cells, extract their features, and hash them for real-time comparison with the training database. Each cell is assigned with multiple weights or probability scores after this matching process. Finally, the classification of the testing image is achieved by aggregating the probability scores of all its cells. In the following sections, we introduce the details of the employed hashing method and our proposed strategy for cell-level analysis.
2.2 Kernelized and Supervised Hashing for Large-Scale Image Retrieval
In this section, we briefly introduce the hashing method employed as our baseline. For each segmented cell, two features are extracted, i.e., GIST [25] and HOG [26], and both of which are hundreds of dimensions, causing issues for the computational efficiency of comparing all samples. To this end, hashing methods have been widely used to compress features into binary codes with merely tens of bits. As a result, such short binary features allow mapping into a hash table for efficient retrieval, e.g., constant-time. To improve the accuracy, the kernelized scheme [18] is usually utilized to handle practical data that is mostly linearly inseparable:
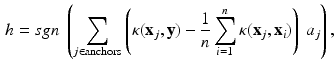
where 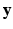 is the feature (e.g., GIST or HOG) to be compressed as binary code,
is the feature (e.g., GIST or HOG) to be compressed as binary code, 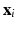 with i from 1 to n means all training samples, i.e., cell patches,
with i from 1 to n means all training samples, i.e., cell patches, 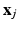 denotes the anchors, i.e., random samples selected from the data, h is the kernelized hashing method taking the sign value of a kernel function with kernel
denotes the anchors, i.e., random samples selected from the data, h is the kernelized hashing method taking the sign value of a kernel function with kernel 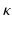 , and
, and 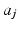 is the coefficient determining hash functions. The resulting binary codes can be used for indexing and differentiating different categories. Although kernelized scheme well solves the linear inseparability problem of features, it is still not able to provide accurate retrieval or classification of cell images, because of their large variations. Therefore, supervised information [20] can be leveraged to bridge the semantic gap by designing more discriminative hash functions:
is the coefficient determining hash functions. The resulting binary codes can be used for indexing and differentiating different categories. Although kernelized scheme well solves the linear inseparability problem of features, it is still not able to provide accurate retrieval or classification of cell images, because of their large variations. Therefore, supervised information [20] can be leveraged to bridge the semantic gap by designing more discriminative hash functions:
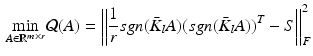
where S is a matrix encoding the supervised information (e.g., 1 for same category and 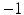 for different categories, which is applicable to multi-class problems) and A is the model parameter to compute hashing code, and
for different categories, which is applicable to multi-class problems) and A is the model parameter to compute hashing code, and ![$$\bar{K}_l = [\mathbf {\bar{k}}(\mathbf {x}_1),\cdots ,\mathbf {\bar{k}}(\mathbf {x}_l)]^T \in \mathbb {R}^{l\times m}$$](/wp-content/uploads/2016/09/A339424_1_En_23_Chapter_IEq9.gif) is the matrix form of the kernel function, in which
is the matrix form of the kernel function, in which 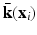 is a kernelized vectorial map
is a kernelized vectorial map 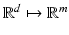 ,
, ![$$A = [\mathbf {a}_1,\cdots ,\mathbf {a}_r] \in \mathbb {R}^{m \times r}$$](/wp-content/uploads/2016/09/A339424_1_En_23_Chapter_IEq12.gif) . The optimization of
. The optimization of  is based on Spectral Relaxation [27] for convexification, which is used as a warm start, and Sigmoid Smoothing that applies standard gradient descent technique for accurate hashing.
is based on Spectral Relaxation [27] for convexification, which is used as a warm start, and Sigmoid Smoothing that applies standard gradient descent technique for accurate hashing.
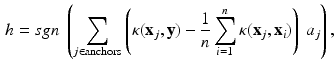
(1)
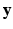 is the feature (e.g., GIST or HOG) to be compressed as binary code,
is the feature (e.g., GIST or HOG) to be compressed as binary code, 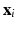 with i from 1 to n means all training samples, i.e., cell patches,
with i from 1 to n means all training samples, i.e., cell patches, 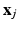 denotes the anchors, i.e., random samples selected from the data, h is the kernelized hashing method taking the sign value of a kernel function with kernel
denotes the anchors, i.e., random samples selected from the data, h is the kernelized hashing method taking the sign value of a kernel function with kernel 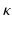 , and
, and 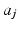 is the coefficient determining hash functions. The resulting binary codes can be used for indexing and differentiating different categories. Although kernelized scheme well solves the linear inseparability problem of features, it is still not able to provide accurate retrieval or classification of cell images, because of their large variations. Therefore, supervised information [20] can be leveraged to bridge the semantic gap by designing more discriminative hash functions:
is the coefficient determining hash functions. The resulting binary codes can be used for indexing and differentiating different categories. Although kernelized scheme well solves the linear inseparability problem of features, it is still not able to provide accurate retrieval or classification of cell images, because of their large variations. Therefore, supervised information [20] can be leveraged to bridge the semantic gap by designing more discriminative hash functions: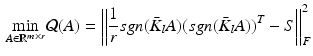
(2)
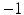 for different categories, which is applicable to multi-class problems) and A is the model parameter to compute hashing code, and
for different categories, which is applicable to multi-class problems) and A is the model parameter to compute hashing code, and ![$$\bar{K}_l = [\mathbf {\bar{k}}(\mathbf {x}_1),\cdots ,\mathbf {\bar{k}}(\mathbf {x}_l)]^T \in \mathbb {R}^{l\times m}$$](/wp-content/uploads/2016/09/A339424_1_En_23_Chapter_IEq9.gif) is the matrix form of the kernel function, in which
is the matrix form of the kernel function, in which 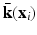 is a kernelized vectorial map
is a kernelized vectorial map 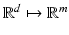 ,
, ![$$A = [\mathbf {a}_1,\cdots ,\mathbf {a}_r] \in \mathbb {R}^{m \times r}$$](/wp-content/uploads/2016/09/A339424_1_En_23_Chapter_IEq12.gif) . The optimization of
. The optimization of 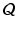 is based on Spectral Relaxation [27] for convexification, which is used as a warm start, and Sigmoid Smoothing that applies standard gradient descent technique for accurate hashing.
is based on Spectral Relaxation [27] for convexification, which is used as a warm start, and Sigmoid Smoothing that applies standard gradient descent technique for accurate hashing.Indexing these compressed features in a hash table, our framework can match each cell of the testing image with all cells in the training database in constant-time. The category of each cell is decided straightforwardly with the majority logic of retrieved cells, and the whole image is hence classified by aggregating results of all cells from the testing image. The whole process is very efficient and takes 1–2 s.
2.3 Weighted Hashing with Multiple Cues
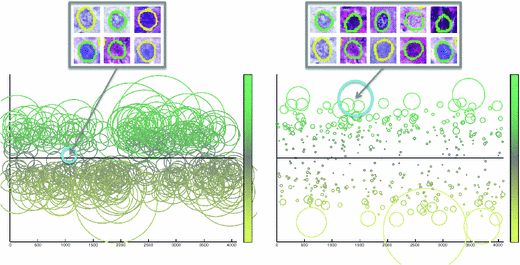
Fig. 2.
We visualize hash tables and their entries according to cells mapped into them, and each circle represents one entry. The left hash table corresponds to HOG feature, and the right is GIST. The x-axis represents different hash entries corresponding to 12 bits, indicating 4096 different entries. The y-axis means the ratio between the adenocarcinoma and squamous carcinoma, ranging from 0 to 1, which is also visualized as different colors. The size of each circle denotes the number of cells mapped into that entry. As shown in the figures, different features may result in diverse cell distributions in the hash table, making it essential to explore consistent measures for feature fusion.
Despite its efficacy in large-scale image retrieval, KSH still has several limitations when dealing with our use case, which requires to hash a large number of cell images. First of all, it builds hash functions for one type of feature, while it is preferred to model multiple cues of cells for accurate classification. Second, multiple cells can be mapped into the same hash entry using KSH, i.e., the hamming distances among them are zero. In this case, one may use majority voting to decide the label of a testing cell image having the same hash entry. However, cell images from different categories can be easily mapped into the same hash entry, due to image noise, erroneous segmentation results, and the low inter-class variations. In other words, not all hash entries are reliable for classification. Figure 2 visualizes the hash tables of two features, GIST [25] and HOG [26], representing the texture characteristics of cells. The entries in each hash table are illustrated according to the distribution of cells mapped into them, such as the ratio between two categories and the number of cells mapped into that entry. The indecisive hash entries are usually around the 0.5 ratio, indicating equal opportunity for either category. The small circles in Fig. 2 are also not reliable, since only few cells are mapped there, which can be easily affected by the image noise or erroneous segmentation. A potential solution is to identify reliable hash entries and omit indecisive one, by heuristically select or prune them via feature selection. However, this may involve tuning parameters and have difficulties in modeling multiple cues of cells because of lacking the consistent measures. Furthermore, it is hard to guarantee that the selected hash entry is sufficiently discriminative for classification. Therefore, we introduce a unified formulation to solve these problems in a principled way. First, probability scores are assigned to each hash entry, based on its ability to differentiate different categories. Then, such probability scores can be integrated from different types of features, by emphasizing reliable hash entries of certain features. In this section, we introduce the details of our method.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree