Molecular features are now essential in distinguishing between glioma histologic subtypes. Currently, isocitrate dehydrogenase mutation, 1p19q codeletion, and MGMT methylation status play significant roles in optimizing medical and surgical treatment. Noninvasive pretreatment and post-treatment determination of glioma subtype is of great interest. Although imaging cannot replace the genetic panel at present, image findings have shown promising signs to identify and diagnose the types and subtypes of gliomas. This article details key imaging findings in the most common molecular glioma subtypes and highlights recent advances in imaging technologies to differentiate these lesions noninvasively.
Key points
- •
Molecular features are necessary for diagnosis of glioma subtype and greatly impact prognosis and treatment response.
- •
Increased contrast enhancement, sharp tumor margins, and homogenous signal intensity are characteristic of IDH-mutant gliomas, whereas degree of necrosis is not.
- •
T2-FLAIR mismatch is specific to IDH-mutant, 1p19q nondeleted gliomas (diffuse astrocytoma).
- •
Advanced MR imaging techniques, such as sodium MR imaging, deep learning, diffusion kurtosis imaging, and texture analysis, are used to predict molecular subtypes.
Background
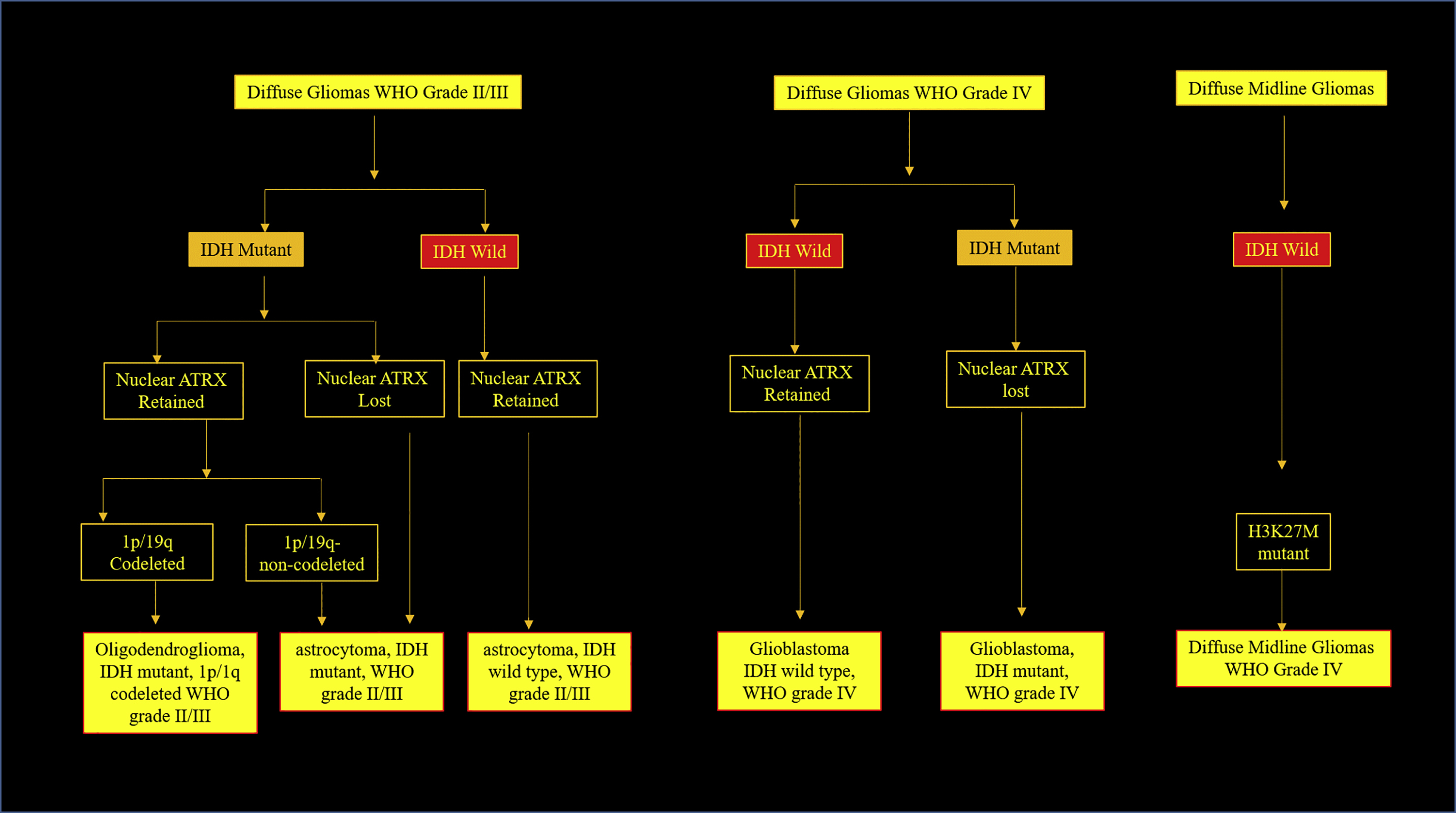
The 2016 World Health Organization (WHO) Classification of Tumors of the Central Nervous System set forth a revised classification system for brain tumors. This update included molecular aberrations in the definition of particular brain tumors and, to some extent, explained why some tumors of identical cell types appear similar on histology but respond differently to the same therapy and have different prognoses. Today, this is thought to be caused by different genetic makeups of tumors, generating tremendous interest in the reclassification of cancers. Several molecular markers including isocitrate dehydrogenase (IDH), 1p/19q codeletion, O6-methylguanine-DNA methyltransferase methylation (MGMT), telomerase reverse transcriptase gene (TERT), α-thalassemia/mental retardation syndrome X-linked gene (ATRX), and p53, were identified as necessary for diagnoses of various gliomas ( Fig. 1 ).
There is parallel effort from an imaging perspective to be able to obtain molecular and genetic information from structural and molecular imaging techniques to match the new genotypic classifications. A major initiative to accomplish this was The Cancer Genome Atlas, undertaken by the National Institutes of Health. This led to the Cancer Imaging Program, which obtains radiologic imaging data for The Cancer Genome Atlas patients and makes it available via The Cancer Imaging Archive. The first large-scale imaging genomic study performed in glioblastomas (GBM) paved the way for the potential correlation between imaging features and histologic patterns and genetic profiles of the tumor, known collectively as “radiohistogenomic interpretation.” Radiomic-imaging features play an important role in accurate and early prediction of tumor genotype when genetic sequencing is not available.
Genetic factors
The 2016 central nervous system (CNS) WHO classification offered an attempt to standardize the terminology combining histopathologic and molecular features. Today CNS tumor naming includes histologic appearance (ie, astrocytoma or oligodendroglioma), WHO grade (II, III), and genetic features. Work on genotyping and identifying specific markers for brain tumors is ongoing. Histologic appearance is decided by routine staining techniques used in histopathology, which decides the cell lineage for the tumor. WHO grading (WHO I, II, III, and IV) of the diffuse glioma depends on cellularity, cortical infiltration, nuclear pleomorphism, hyperchromasia, and mitotic figures. The new classification posits grade I gliomas as virtually nonexistent in adults. Currently, the main molecular makers of diagnostic significance for gliomas include IDH, 1p19q deletion, MGMT, TERT, ATRX, and a tumor protein p53 gene (TP53). ,
Diffuse gliomas based on gene expression and available molecular markers are classified as follows: grade II (diffuse astrocytoma, IDH mutant) and grade III astrocytic tumors (anaplastic astrocytoma, IDH mutant), grade II (oligodendroglioma IDH mutant and 1p/19q codeleted) and grade III oligodendrogliomas (anaplastic oligodendroglioma IDH mutant and 1p/19q codeleted), grade IV GBM (GBM IDH mutant and IDH wild-type), and diffuse gliomas of childhood (diffuse midline glioma, H3-K27M-mutant) (see Fig. 1 ). , Whenever molecular diagnostic testing is not available or genetic assay testing is inconclusive, the Not Otherwise Specified (NOS) category is used.
Neuro-oncology teams are of the opinion that much work still needs to be done to have a clear understanding of the genetic basis for many of the other brain tumors, including GBM. Following the 2016 classification, the Consortium to Inform Molecular and Practical Approaches to CNS Tumor Taxonomy (cIMPACT-NOW) was created in late 2016 under the sponsorship of the International Society of Neuropathology to provide a forum to evaluate and recommend proposed changes to future CNS tumor classifications. cIMPACT-NOW updates are intended to provide guidance for diagnosticians and potentially inform future WHO classifications. This article focuses on correlating the phenotypic and genotypic features of gliomas, gliomagenesis of GBM, IDH mutation, other molecular markers and mutations in diffuse gliomas, and their imaging correlates on various MR imaging techniques.
Isocitrate Dehydrogenase Status
In humans, IDH exists in three isoforms (IDH1, IDH2, and IDH3). IDH1 and IDH2 are proteins in the cytosol and mitochondria, respectively, that generate reduced nicotinamide adenine dinucleotide phosphate from NADP + by catalyzing the oxidative decarboxylation of isocitrate to α-ketoglutarate outside of the Krebs cycle ( Fig. 2 ). IDH3 functions to convert isocitrate to α-ketoglutarate and NAD + to NADH in the Krebs cycle. IDH plays an important role in cellular defense against oxidative stress. Cells with low levels of IDH become more sensitive to oxidative damage. IDH1 mutations involve an amino acid substitution (glycine to arginine) in the active site of the enzyme in codon 132 (R132H). This mutation results in the abnormal production of 2-hydroxyglutarate (2HG), which causes histone and DNA methylation, thereby promoting tumorigenesis. IDH2 mutations occur in codon 172 and are associated with 2-hydroxyglutaric aciduria. These result in seizures, weak muscle tone (hypotonia), and progressive damage to the brain parenchyma. Inhibition of these enzymes results in widespread histone and DNA methylation, which in turn leads to increased tumorigenesis. Mutant IDH1 and IDH2 occur in more than 70% of WHO grade II and III astrocytomas, oligodendrogliomas, and in secondary GBM that developed from the previously mentioned lower-grade lesions.
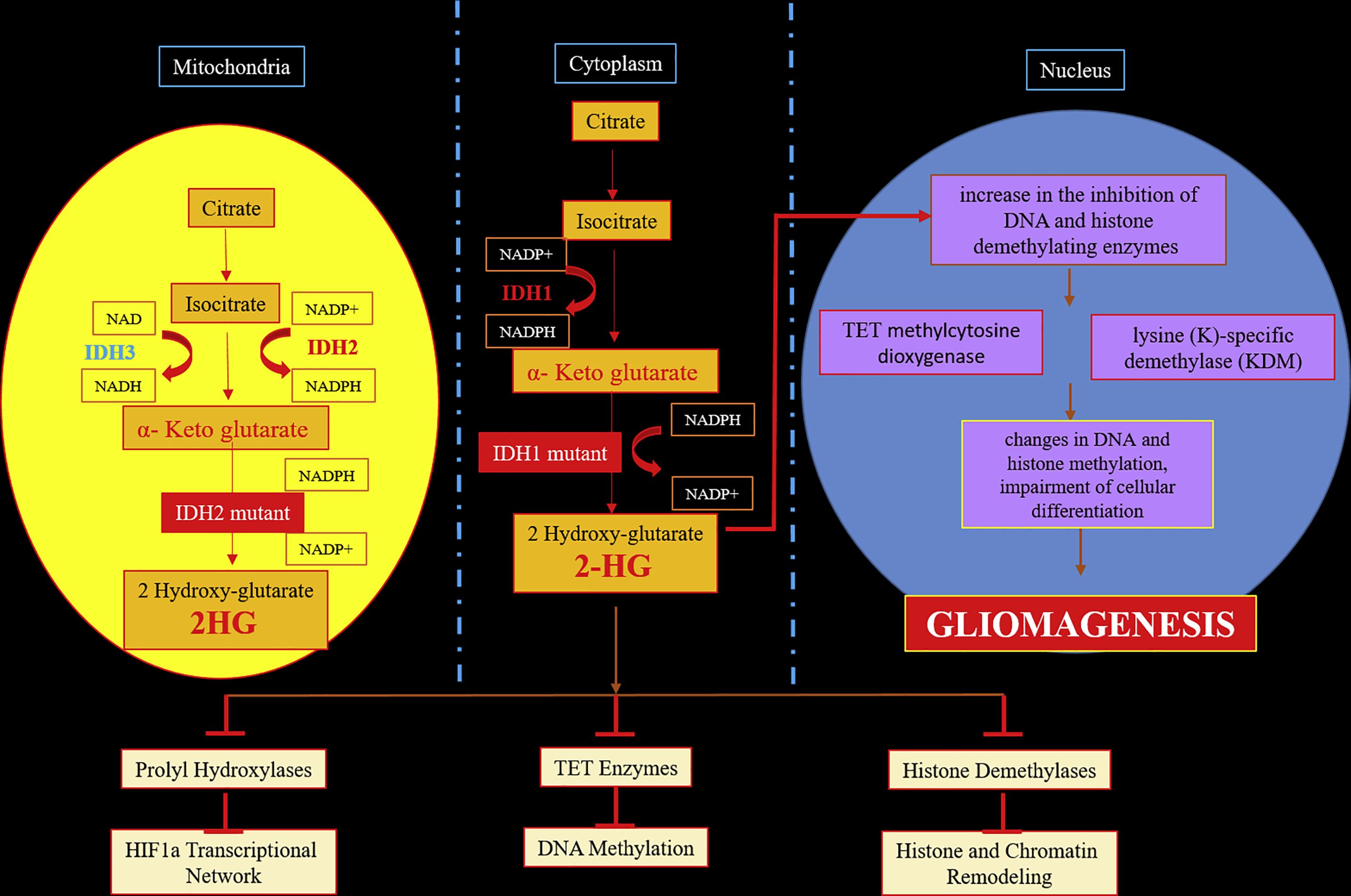
IDH1 and IDH2 are enzymes that function at the crossroads of cellular metabolism, epigenetic regulation, redox states, and DNA repair. The pro-oncogenic effect of IDH mutations is caused by the damage produced by high levels of reactive oxygen species to DNA and by 2HG, an oncometabolite that alters cell proliferation. , 2HG impairs the maturation of extracellular collagen in the brain capillary network basement membrane, which promotes cell migration to the extravascular space and intravascular fluid leakage into the extracellular space. This leakage is reflected as enhancement on contrast-enhanced MR imaging and permeability on MR perfusion imaging.
The Cancer Genome Atlas Research Network found an IDH mutation rate of 80% of grade II/III gliomas (diffuse astrocytomas, oligodendrogliomas, and oligoastrocytomas) and only 10% in GBM. , Based on these results, the WHO now recognizes the IDH mutation as a critical biomarker in glioma classification. Among the GBM, IDH1/IDH2 mutations are more commonly seen in secondary GBM originating from lower-grade diffuse gliomas than in primary GBM. When both IDH1/IDH2 mutant are negative, as in the primary GBM, it is labeled as IDH wild-type. If IDH testing is not available, cannot be fully performed, or is inconclusive, the GBM is labeled IDH NOS. IDH status is considered an independent determinant of prognosis in patients with diffuse gliomas. Survival is higher in IDH -mutant patients than in IDH wild-type patients.
MGMT Methylation
MGMT is an apoptotic enzyme and DNA repair protein that inhibits the cross-linkage of double-stranded DNA, and repairs premutagenic, precarcinogenic, and pretoxic DNA damage. Methylation within the MGMT gene promoter on chromosome 10q26.3 induces loss of function of the protein, ultimately leading to insufficient DNA repair and tumorigenesis. , For brain tumors, MGMT activity declines as glioma grade increases.
Alkylating chemotherapeutics (ie, methylating agents [procarbazine, dacarbazine streptozotocin, and temozolomide] and chloroethylating agents [carmustine, lomustine, nimustine, and fotemustine]) are commonly used in the treatment of brain tumors, malignant melanoma, and lymphoma because they are powerful inducers of apoptosis. These alkylating agents cause mutations, sister chromatid exchanges, recombination and chromosomal aberrations, and DNA mismatch repair to DNA double-strand breaks that trigger cell death. However, their efficacy largely depends on MGMT expression. The loss of MGMT protein expression caused by MGMT promoter methylation reduces the DNA repair activity of glioma cells, preventing their resistance to alkylating agents. Patients with GBM and a methylated MGMT promoter are more sensitive to the killing effects of alkylating drugs because tumor cells with low MGMT expression are unable to repair such DNA lesions and are prone to apoptosis. Cancer cells that overexpress MGMT are resistant to alkylating agents. Therefore, MGMT promoter methylation is the most relevant prognostic marker and is used to predict a therapeutic response to alkylating agents. Documentation of MGMT promoter methylation is important to decide the choice of chemotherapy or radiotherapy in patients with brain tumors.
Methylation of the promoter region of the MGMT gene is more frequently found in secondary GBMs compared with primary GBMs (75% vs 36%). , By knowing whether or not the MGMT promoter is methylated, response to temozolomide, a standard adjuvant chemotherapy treatment of GBMs, may be predicted. Overall survival (OS) in methylated patients is better when temozolomide is given concurrently with radiation therapy. In unmethylated patients, radiotherapy alone is more effective. , , In addition, GBM patients with MGMT promoter methylation have better OS and progression-free survival (PFS) than those without methylated MGMT promoters regardless of therapeutic intervention. MGMT promoter methylation is also a strong predictor of pseudoprogression and IDH mutation to the extent that almost all patients with an IDH mutation also exhibit MGMT promoter methylation.
1p/19q Codeletion
According to the 2016 WHO classification of CNS tumors, 1p19q codeletion is required for the diagnosis of oligodendroglioma. A 1p/19q codeletion is the complete deletion of the short arm of chromosome 1 (1p) and the long arm of chromosome 19 (19q). , It is a definitive marker for grades II and III (anaplastic) oligodendroglioma. In contrast, a patient that is IDH positive and lacks a 1p19q codeletion carries a diagnosis of a diffuse astrocytoma (see Fig. 1 ). ,
It is important to remember that oligodendroglial tumors have neither ATRX nor TP53 gene mutations. An oligodendroglioma that is IDH-mutant but has not been analyzed for 1p/19q status is designated as oligodendroglioma NOS, whereas an oligodendroglioma that is IDH-mutant with a 1p deletion but an intact 19q is designated as diffuse glioma, IDH-mutant with 1p loss/19q retention, Not Elsewhere Classified (NEC). , , , Histopathologically diagnosed oligodendrogliomas without 1p19q codeletion is classified as a diffuse glioma of the oligodendroglial phenotype. Pediatric oligodendrogliomas are IDH negative, do not display 1p/19q codeletion, and are classified as NOS.
The presence of 1p/19q codeletion is associated with a favorable response to chemotherapy and radiotherapy and ultimately patient prognosis and survival. , , 1p/19q codeletion is also linked to sensitivity to procarbazine-lomustine-vincristine chemotherapy and improved outcomes in patients with oligodendroglioma. ,
Molecular genetics of glioblastoma and gliomagenesis
Gliomas are CNS tumors of glial origin, and GBM are the most common and aggressive subtype. On the phenotyping and genotyping features, GBM is classified into primary (95%) and secondary GBMs (5%). Primary GBMs are aggressive, highly invasive, and more commonly seen in the elderly. , , , Secondary GBMs are much less common, arise from low-grade gliomas, mostly seen younger than the age of 45, and have a better prognosis. GBMs are histologically indistinguishable but show distinctive genetic alterations that allows differentiation.
Genetic alterations occur in major key pathways to form GBMs. Gliomagenesis is a multifactorial process involving several genetic mutations. The introduction of genotyping has had a major impact on the classification, treatment, and understanding of outcome for GBMs. IDH status classifies GBMs into three types: IDH wild-type, IDH-mutant, and IDH NOS. , , ,
- 1.
IDH wild-type GBMs are also called primary or de novo GBMs. They form around 95% of the total GBMs and are primarily seen in patients older than 55. In contrast to the IDH-mutant, IDH wild-type follow an aggressive course and have poor prognosis. They are associated with various genetic alterations including epidermal growth factor receptor (EGFR), MGMT, the phosphatase and tensin homolog, TP53 , platelet-derived growth factor receptor-α (PDGFRA), neurofibromin 1 (NF1), cyclin-dependent kinase inhibitor 2A and B (CDKN2A/B) genes, and telomerase reverse transcriptase (TERT) promoter.
The most commonly pathway involved is receptor tyrosine kinases, which bind with growth factors inducing a conformational shift. This shift activates the kinase function of the receptor tyrosine kinases allowing cross-phosphorylation of tyrosine residues in preparation for downstream signaling cascades. EGFR functions in the proliferation, migration, differentiation, and survival of all types of CNS cells. , In GBM cells, EGFR signaling gets activated either through overexpression of the receptor and its ligand amplifying the EGFR function . Amplification of the EGFR gene and activating mutations of its protein product are hallmarks of primary GBM and are associated with TP53 mutations. , , , PTEN amplification and loss of chromosome 10 are additional features of primary GBMs. Verhaak and colleagues described a robust gene expression–based molecular classification of GBMs into proneural, neural, classical, and mesenchymal subtypes that integrate multidimensional genomic data to establish patterns of somatic mutations and DNA copy number. Primary GBMs represent the classical, mesenchymal, and neural subtypes. The mesenchymal and classical subtypes are typically associated with more aggressive, higher-grade gliomas. The classical subtype demonstrates a greater preponderance of EGFR amplification, decreased rates of TP53 mutation, and p16INK4A and p14ARF deletion.
- 2.
IDH-mutant GBMs, also called secondary GBMs, usually arise from diffuse WHO grade II or III gliomas. Genetic alterations common to secondary GBMs include TP53 mutations and IDH1/2 mutations. , , TP53 mutations are detectable in the early stages of secondary GBM. IDH1/2 mutations rarely occur in primary GBMs and are therefore the most reliable indicator to differentiate between primary and secondary GBMs. Secondary GBMs are also associated ATRX (80%) and retinoblastoma protein 1 (Rb1) mutations, reflecting its origin from lower-grade gliomas. , It is primarily seen within the frontal lobe and corresponds to the proneural histologic subtype on the Verhaak and colleagues classification. The proneural subtype is less aggressive; seen in younger patients; and has alterations in TP53, PDGFRA, PIK3C, and IDH1
- 3.
The NOS group includes tumors for which the IDH status cannot be determined. , Another recently introduced category is NEC, used when diagnostic testing has been successfully performed, but the results do not allow for a WHO categorization. In contrast, the NOS designation is used when a full molecular work-up has not been undertaken or was not successful. NEC diagnoses are descriptive diagnoses where the pathologist uses a non-WHO term to describe the tumor.
Tumor heterogeneity, one of the hallmarks of GBM, is the presence of multiple different cell subpopulations within a single tumor. It is caused by cancer stem cells that possess varying degrees of stemness, the ability to self-renew and differentiate into different tumor cell types, and clonal evolution that may enhance genetic diversity within the affected tissues. , This heterogeneity varies from different zones of the GBM, namely the core, interface, and peripheral brain zones. Because tumor fragments from the same patient may have different molecular subtypes in different zones, GBM grading is complex. GBM also shows epithelial to mesenchymal transitions, thought to be caused by signaling pathways of Wnt, transforming growth factor-β, and NOTCH. , This transition is partly responsible for the migration, diffusion invasion, and angiogenesis of GBM. By virtue of the inherent heterogeneity of these tumors, not all of the cells within a glioma respond to chemotherapy and radiation, resulting in tumor progression/recurrence.
Imaging
Phenotyping and genotyping using tumor tissue remains the gold standard for characterizing the histologic type and genetic make of the tumor; it is thereby the predominant factor in deciding choice the treatment and prognosis. However, pathology and genotyping have their own limitations. One major downside of pathologic analysis is intratumoral heterogeneity, even across molecular subtypes. It is often not feasible to study every section of invasive tumor, and the availability of the newer diagnostic tools, such as immunohistochemistry, genotyping, and molecular markers, is limited and expensive. Another clinical limitation of genotyping is that the study of a tumor’s genetic profile can only be done on tumor tissue samples processed in the primary institution or laboratory. Furthermore, the usefulness of these techniques is limited once the patient is on radiation or chemotherapy. Unfortunately, there is still no worldwide standard for processing and analyzing tissue samples. The quest for developing a noninvasive, cheaper, and specific technique to identify the molecular/genomic makeup of tumors is still ongoing.
Role of MR imaging
At present, imaging modalities have limitations in locating the point mutation of a tumor’s genetic material. However, the Cancer Imaging Program and The Cancer Imaging Archive have paved the way to correlate imaging features and histologic patterns with the genetic profile of the tumor; this is called “radiohistogenomic interpretation.” , , Thus, in the absence of genetic sequencing, imaging modalities play an important role.
Molecular imaging using MR imaging techniques, such as diffusion-weighted imaging/apparent diffusion coefficient (ADC), MR spectroscopy, MR perfusion, dynamic susceptibility contrast MR perfusion, and diffusion tensor imaging has shown promising results in understanding the genetic profile and biologic behavior of the tumor. Both structural and molecular imaging play an important role in the radiohistogenomic classification of the brain tumor. Markers that have been found useful in these classifications include location of the tumor, ADC values, FLAIR/T2 hyperintensities, chemical analysis of the tumor mass and surrounding brain using MR spectroscopy, and texture analysis using a combination of the previously mentioned techniques. These techniques are widely used to diagnose the IDH status; MGMT methylation; and, to a lesser extent, 1p/19q codeletion. , , , Extensive radiogenomic research is in progress to find imaging correlates of the other new molecular markers, such as TERT promoters, ATRX, TP53 mutations, TP53, diffuse midline gliomas, H3 K27M-mutant, and wingless and sonic hedgehog activation. , , , , A noninvasive prediction of IDH mutation is important because maximal surgical resection, including enhancing and nonenhancing tumors, may contribute to a better prognosis in IDH1-mutant gliomas. Although a survival benefit was noted in the complete resection of enhanced IDH1 wild-type gliomas, no survival benefit was observed in further resection of the nonenhanced portion.
Key imaging findings
MGMT Methylation
Standard methods of imaging are limited in their ability to distinguish molecular subtypes of gliomas. These imaging modalities have not been able to clearly differentiate MGMT methylated from unmethylated tumors. Several investigations have evaluated the value of ADC for predicting MGMT promoter methylation, but the results are inconclusive. , However, a minimum ADC value may have some prognostic value in preoperatively estimating the status of MGMT promoter methylation (84% sensitive, 91% specific). If a relationship were to be found between ADC and MGMT promoter methylation, this would be especially useful in noninvasively predicting methylation because accurate measurement of methylation is difficult because of small biopsy specimens and regional heterogeneity of GBM.
Knowing that MGMT promoter methylation has been used as a prognostic biomarker, researchers have also addressed the role of MGMT methylation via imaging in GBMs. Using various MR imaging techniques, it was noted that the diagnostic performance of MR imaging for prediction of MGMT promoter methylation with recently diagnosed GBM patients was clinically viable. This study found that MGMT promoter methylation in GBMs shows less edema, high ADC, and low perfusion on MR imaging, with a sensitivity at 79% (95% confidence interval, 72%–85%), a specificity of 78% (95% confidence interval, 71%–84%), and the area under the receiver operating curve (AUC) at 0.86 (95% confidence interval, 0.82–0.88).
Another study evaluating MGMT promoter methylation in primary GBMs through imaging looked at the relationship between ADC and relative cerebral blood flow (rCBF) values in a manually drawn region of interest. Using a combination of tumor location, necrosis, ADC, and rCBF, the highest AUC resulted in 0.914. This suggests that ADC and rCBF, when used with other known factors of GBMs, are associated with the prediction of MGMT promoter methylation.
Isocitrate Dehydrogenase Mutation
Determination of IDH mutation status is important and is usually determined via polymerase chain reaction, gene sequencing, and immunohistochemistry. However, recent advances in imaging have afforded the potential to define the IDH status of diffuse gliomas ( Table 1 ). , Radiogenomic texture analysis may show that a tumor’s molecular differences and biologic behavior are mirrored in imaging features and that these parameters can be used for patient stratification to optimize glioma treatment. , This classification is clinically significant because IDH wild-type tumors have worse prognoses compared with secondary IDH-mutant tumors or others with more aggressive biologic behavior. The following is a compiled list of imaging findings correlated with IDH status:
- •
IDH-mutant tumors are primarily located in the frontal lobe or subventricular region of the frontal horns of the lateral ventricles and are less likely to invade eloquent areas of the brain. , , , IDH-mutant tumors are rare in the occipital lobe. IDH-mutant gliomas tend to be larger, exhibit slower growth, and have better defined contours than IDH wild-type gliomas. ,
- •
Gliomas with IDH wild-type usually are multilobar; cross the midline; involve the corpus callosum; involve more than one lobe; and commonly affect eloquent areas and deeper structures, such as the diencephalon and brainstem. ,
- •
IDH wild-type GBMs associated with unmethylated MGMT gene promoters are located predominantly in the right hemisphere and have poor prognoses. IDH-mutant MGMT methylated GBMs show better prognosis with temozolomide are located predominantly in the left hemisphere. ,
- •
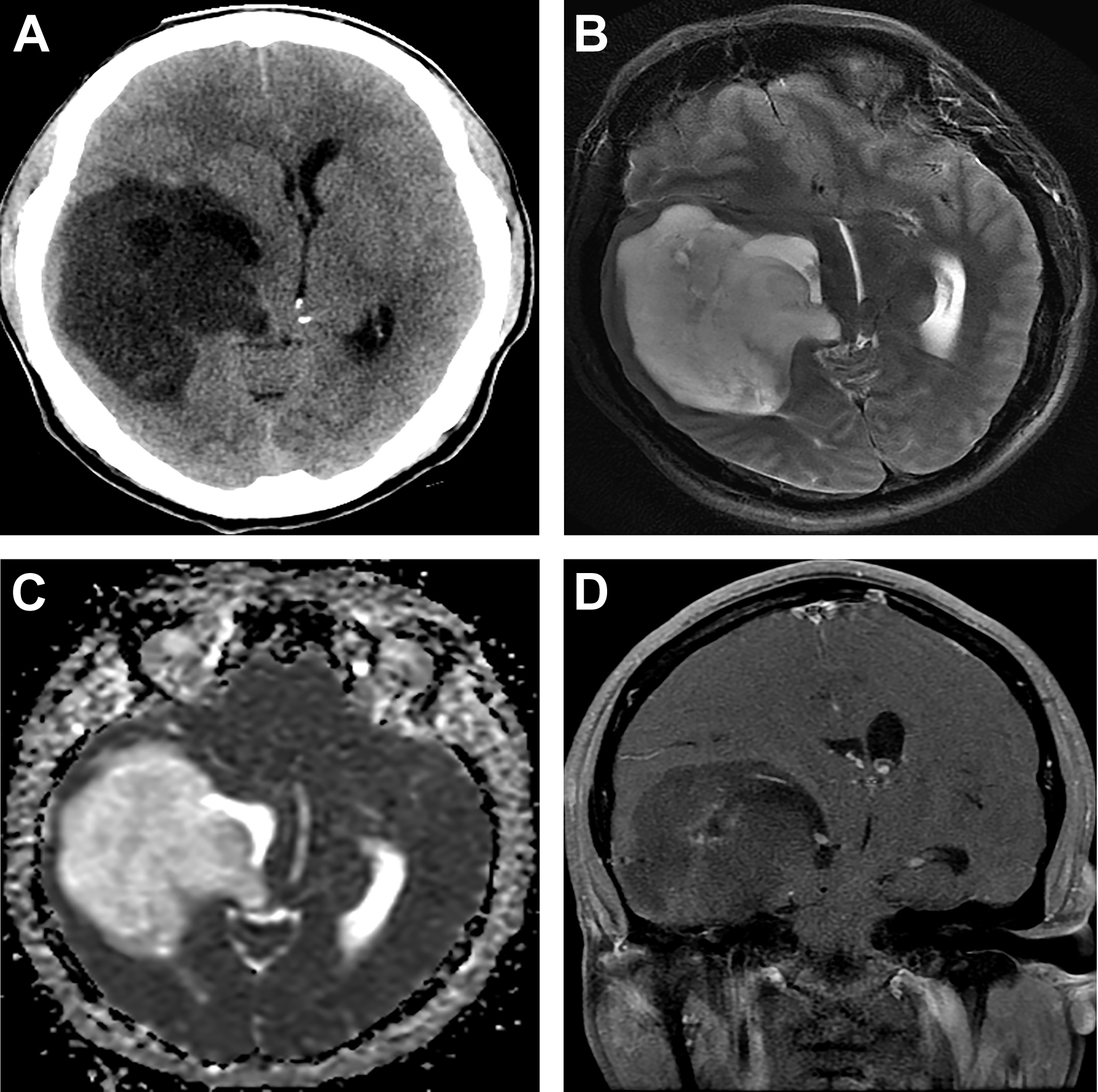
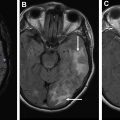
The presence of large portions of nonenhanced tumor in GBMs is strongly associated with the IDH1 mutation ( Fig. 3 ). Large nonenhanced portions are caused by low VEGF levels in IDH-mutant tumors. IDH wild-type GBMs usually show smaller nonenhancing component.