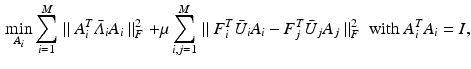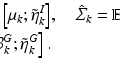consists of N voxels 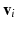 observed at T time points. The functional architecture of a brain state can be modeled as a connectivity graph G with vertices V representing the voxels, and edges E between these vertices. Let W denote the affinity matrix assigning weights to the egdes, where
observed at T time points. The functional architecture of a brain state can be modeled as a connectivity graph G with vertices V representing the voxels, and edges E between these vertices. Let W denote the affinity matrix assigning weights to the egdes, where 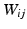 is the Pearson correlation coefficient between the time-series of vertex i and j. The spectral representation of the functional connectivity structure can be established via eigendecomposition of the normalized Laplacian matrix L, defined as
is the Pearson correlation coefficient between the time-series of vertex i and j. The spectral representation of the functional connectivity structure can be established via eigendecomposition of the normalized Laplacian matrix L, defined as 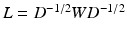 , where D is the diagonal matrix of node degrees, i.e.
, where D is the diagonal matrix of node degrees, i.e. 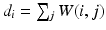 . The normalized Laplacian is a symmetric version of the random walk matrix
. The normalized Laplacian is a symmetric version of the random walk matrix 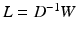 , which is scaling invariant, and can be viewed as a diffusion process, where the transition probabilities for a diffusion time t between nodes can be expressed by
, which is scaling invariant, and can be viewed as a diffusion process, where the transition probabilities for a diffusion time t between nodes can be expressed by 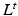 . Eigendecompostion of L results in eigenvalues
. Eigendecompostion of L results in eigenvalues 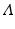 and corresponding eigenvectors
and corresponding eigenvectors 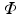 , where
, where 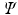 , where each vertex i in the original graph is represented by
, where each vertex i in the original graph is represented by
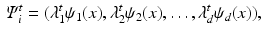
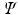 , where the functional relations of the fMRI signals are captured by the diffusion distance
, where the functional relations of the fMRI signals are captured by the diffusion distance 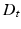 [15]. The diffusion distance
[15]. The diffusion distance 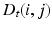 is defined as the probability of traveling from node i to node j in t steps, by considering all possible paths between these two nodes. The Euclidean distance in this new embedding space approximates the diffusion distance
is defined as the probability of traveling from node i to node j in t steps, by considering all possible paths between these two nodes. The Euclidean distance in this new embedding space approximates the diffusion distance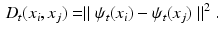
2.2 Initial Correspondence Estimates Across Individuals
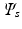 and template embedding
and template embedding 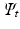 , an orthonormal transformation matrix
, an orthonormal transformation matrix 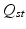 is calculated, accounting for rotations, sign flips and reordering of the spectral dimensions.
is calculated, accounting for rotations, sign flips and reordering of the spectral dimensions. 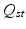 is defined as
is defined as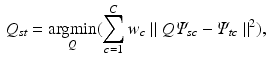
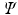 are the embeddings of s and t and w is the Euclidean distance between a pair c of corresponding voxels. After this initial alignment of embeddings, we perform nonrigid point cloud registration to refine the alignment and close potential gaps between the embedded sampling point distributions by Coherent Point Drift [19].
are the embeddings of s and t and w is the Euclidean distance between a pair c of corresponding voxels. After this initial alignment of embeddings, we perform nonrigid point cloud registration to refine the alignment and close potential gaps between the embedded sampling point distributions by Coherent Point Drift [19].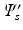 and
and 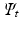 , resulting in n correspondence pairs
, resulting in n correspondence pairs 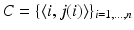 with
with 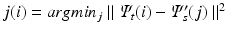 . For each pair of embeddings, a subset of q correspondence pairs C is selected based on their distance from the origin and used as initial coupling for joint diagonalization, described in the following section. Note that the purpose of this initial alignment is only to determine a subset of initial correspondence esimates. Sub-sequent joint diagonalization is based on the individual Laplacians and the correspondence pairs.
. For each pair of embeddings, a subset of q correspondence pairs C is selected based on their distance from the origin and used as initial coupling for joint diagonalization, described in the following section. Note that the purpose of this initial alignment is only to determine a subset of initial correspondence esimates. Sub-sequent joint diagonalization is based on the individual Laplacians and the correspondence pairs.2.3 Joint Diagonalization
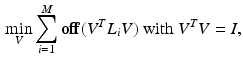
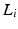 are the dataset specific Laplacians. The off-diagonal penalty
are the dataset specific Laplacians. The off-diagonal penalty 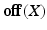 is typically defined as the sum of squared off diagonal elements
is typically defined as the sum of squared off diagonal elements 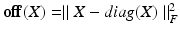 . This problem can be solved with the generalized Jacobi method (JADE method) [2]. It assumes full coupling with an equal amount of samples for every dataset, i.e. every sampling point in a dataset has a corresponding point in all other datasets. In our case we cannot assume identical numbers of sampling points or complete given correspondences between datasets. Instead we only use a sub-set of paired vertices to guide the joint diagonalization.
. This problem can be solved with the generalized Jacobi method (JADE method) [2]. It assumes full coupling with an equal amount of samples for every dataset, i.e. every sampling point in a dataset has a corresponding point in all other datasets. In our case we cannot assume identical numbers of sampling points or complete given correspondences between datasets. Instead we only use a sub-set of paired vertices to guide the joint diagonalization.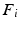 is a
is a 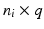 coupling matrix, where
coupling matrix, where 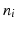 is the number of data points and q is the number of correspondences. Based on this sparse subset, eigenbases
is the number of data points and q is the number of correspondences. Based on this sparse subset, eigenbases 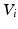 are established, such that
are established, such that 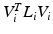 is approximately diagonal and the eigenbases correspond for corresponding samples
is approximately diagonal and the eigenbases correspond for corresponding samples 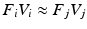 . This results in multiple dataset specific eigenbases, contrary to the original joint diagonalization approach [2].
. This results in multiple dataset specific eigenbases, contrary to the original joint diagonalization approach [2].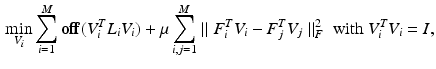
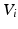 are the dataset specific eigenbases,
are the dataset specific eigenbases, 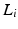 the data specific Laplacians and
the data specific Laplacians and 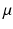 is a coupling weight [6]. The sparse coupling between two datasets i and j is defined by corresponding points
is a coupling weight [6]. The sparse coupling between two datasets i and j is defined by corresponding points 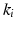 and
and 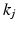 and encoded as
and encoded as 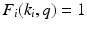 and
and 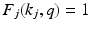 , with zeros elsewhere in the
, with zeros elsewhere in the 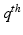 column. However, the coupling is not restricted to such point-wise coupling, and can contain any correspondence weighting. We use the leading k dimensions of the embedding, since they hold the majority of information. This reduces the computational complexity to an optimization problem with
column. However, the coupling is not restricted to such point-wise coupling, and can contain any correspondence weighting. We use the leading k dimensions of the embedding, since they hold the majority of information. This reduces the computational complexity to an optimization problem with 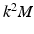 variables, instead of
variables, instead of 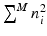 variables for full eigenbases. Then, the formulation in Eq. 5 can be rewritten as
variables for full eigenbases. Then, the formulation in Eq. 5 can be rewritten as