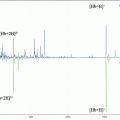
Fig. 1
Cutting apparatus used for longitudinal sectioning of hair samples. (a) Image of the in-house built cutting apparatus and (b) schematic of the cutting apparatus
Following decontamination, place a single hair sample into one of the grooves of the cutting block and fix at one end with a small piece of double-sided tape, as shown in Fig. 2a (see Notes 3 – 5 ).


Fig. 2
Method for preparing longitudinal sections of single hair samples. (a) Hair sample attached to the cutting block with double-sided tape , (b) preparing longitudinal sections of single hair samples using in-house built cutting apparatus, (c) longitudinal sections mounted on conductive glass slide with double-sided copper tape
While holding the other end with a gloved finger, slowly run the cutting device along the length of the hair (Fig. 2b).
Place another piece of double-sided tape on the other end of the hair and transfer onto double-sided copper tape mounted on a ITO glass slide, press the hair into the tape using a clean glass slide (Fig. 2c).
Use a Leica DMRX microscope equipped with a digital camera to determine if the hair section has been successfully transferred onto the glass slide.
3.3 Preparation of Cross-Sections
- 1.
Following decontamination, thread the hair sample through the lid of the tube and using a paper clip, attach two small magnets at the end of the single hair sample. Attach two more magnets at the opposite end of the hair sample.
- 2.
Place the hair sample into a plastic tube that contains a 10% solution (w/v) of gelatin, as shown in Fig. 3b (see Note 6 ).

Fig. 3
Method for embedding single hair samples. (a) Materials required for the embedding of single intact hair samples, (b) single hair sample clipped between magnets and placed in a plastic tube filled with embedding material, (c) embedded hair sample following snap freezing, and (d) cryo-sectioning of embedded hair samples
- 3.
Snap-freeze the contents by placing the plastic tube in liquid nitrogen for 30 s (Fig. 3c).
- 4.
Remove the embedded hair from the plastic tube and cut into 1 cm blocks (the average growth of hair being 1 cm/month), mount one of the blocks onto a cryo-microtome stage using a drop of water.
- 5.
Section the embedded hair samples at −20 °C using a cryo-microtome to produce 12 μm thick sections, thaw mount sections onto a clean indium tin oxide (ITO) glass slide (see Note 7 ).
- 6.
Inspect the sections using a Leica DMRX microscope equipped with a digital camera to determine the position of the hair cross-section and mark the location on the opposite side of the glass slide using a permanent marker pen.
3.4 Matrix Deposition for MALDI-MS/MS Imaging
- 1.
Place the sample in the ImagePrep matrix application device.
- 2.
Fill the bottle with the matrix solution.
- 3.
Select the manufacturer’s method for spraying CHCA and start the sequence.
- 4.
Use a Leica DMRX microscope equipped with a digital camera to inspect the crystal coverage.
3.5 Metal Deposition for MetA-SIMS Imaging
- 1.
Place the sample in the chamber of the Quorum Technologies sputter coater and close the lid.
- 2.
Select the density of the metal (19.30 g/cm3 in the case of gold) and the desired thickness (1 nm) on the film thickness monitor.
- 3.
Set the discharge voltage to 1.5 kV and the plasma current to 25 mA in order to achieve a homogenous coating.
- 4.
Start the sequence (operate in automatic mode), once the sequence is complete vent the instrument and remove the sample (see Note 8 ).
3.6 MALDI-MS/MS Imaging
- 1.
Calibrate the instrument prior to analysis with either a standard mixture of polyethylene glycol (PEG 200-3000) in water mixed with matrix or a saturated solution of red phosphorus in acetone.
- 2.
Spot 0.5 μL of a cocaine base standard (100 ng/μL in 70% MeOH) onto a MALDI target plate, followed by 0.5 μL of the matrix solution.
- 3.
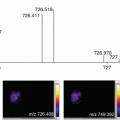
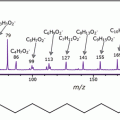
Optimize the instrumental settings using the cocaine base standard such as the laser power and collision energy (trap cell). The optimal settings were as follows: laser power 250 (200 Hz) and collision energy 10 eV (monitor the main product ion of cocaine at m/z 182 formed by neutral loss of benzoic acid).
- 4.
Following method optimization , attach the sample onto a glass slide adaptor using double-sided tape and scan using a flatbed scanner to produce a digital image, ensure the image quality is around 600 dpi or better.
- 5.
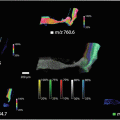
Import the digital image of the sample into the MALDI imaging pattern creator software, to define the area to be imaged and the spatial resolution (150 × 50 μm).
- 6.
Set up the MS/MS imaging method in the MassLynx 4.1 software. The settings were as follows: positive ion mode, V-mode, mass range m/z 50–350, also use the previously optimized settings (see step 3).
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree