Fig. 1
Electron micrographs of silicon-based nanostructure surfaces used in NIMS experiments. A unique feature of these surfaces is that they are UV-absorbing thermal insulators with a large surface area, facilitating their unique desorption/ionization properties
Hydrophobic fluorous materials introduced into nanostructured surfaces have also played an important role in producing surfaces that allow for improved performance for NIMS including enhanced sensitivity (Fig. 2). Three different methods have been developed to incorporate fluorous compounds within porous silicon nanostructures. First, silicon nanostructures have been designed with a covalent pentafluorophenyl modification to reduce analyte adhesion and protect the porous surface from oxidation [5]. A second method has applied the addition of fluorous surfactants, such as perfluoroundecanoic acid, with the pentafluorophenyl-modified silicon surface. These surfaces have been shown to be more effective at reducing analyte adhesion and improving desorption/ionization efficiency [6]. The third method, introduced in 2007, employs fluorous siloxanes as liquid initiators to coat the porous silicon nanostructure surface and further minimize analyte adhesion [7]. With this NIMS technology, it was found that fluorous siloxane initiators did not absorb laser light or ionize, and therefore do not contribute chemical noise in the spectrum, a very important aspect of the NIMS design. Subsequent laser-induced heating transfers energy to the trapped liquid phase, causing rapid initiator vaporization and desorption/ionization of the intact analytes without fragmentation. Among its features is that this surface is stable in ambient air, has an expanded mass range, and can be used to analyze biofluids and image tissues (Fig. 3). The versatility of the fluorinated NIMS platform has now been demonstrated for a large variety of analytes, ranging from metabolites and drugs to peptides and proteins [4–7].
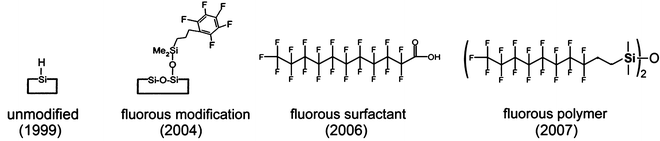
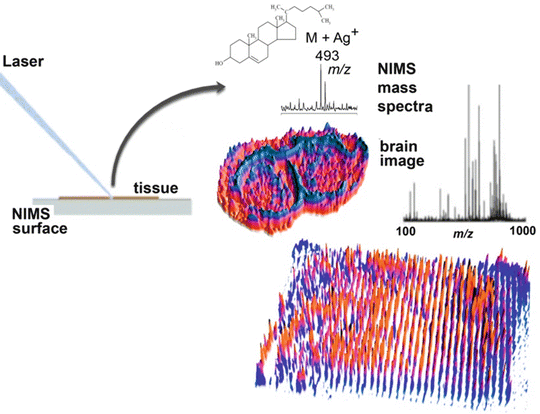

Fig. 2

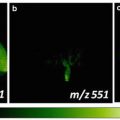
Fig. 3
Nanostructure imaging mass spectrometry (NIMS) of a brain tissue and also imaging of a plate containing 1,500 discrete chemical entities spotted on the NIMS surface
3 Ultrahigh-Sensitivity Detection
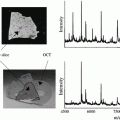
The ultrahigh sensitivity that can be obtained with NIMS has been successfully demonstrated with specific analytes down to the yoctomole level as shown in Fig. 4. The first report of yoctomole sensitivity with NIMS was using a pentafluorophenyl-silylated nanostructure silicon surface to analyze des-Arg9-bradykinin (des-Arg9-bradykinin is commonly used by instrument manufacturers to test sensitivity). Here a series of dilution experiments was carried out to ultimately demonstrate a lower limit of detection for the peptide at 480 molecules (800 ymol) (Fig. 3a) [5]. Similarly, NIMS was also found to have yoctomole detection for small molecules where lower limits of detection of 700 ymol for verapamil [18] and 650 ymol for propafenone have been observed [19] (Fig. 3b). Given the significance of this unprecedented sensitivity, the experiments were replicated on ten separate occasions by three different individuals.
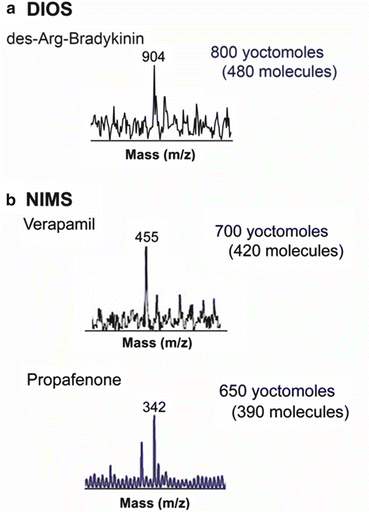

Fig. 4
High-sensitivity nanostructure imaging mass spectrometry (NIMS) experiments. Detection limit of (a) 480 molecules (800 ymol) for des-Arg9-bradykinin using pentafluorophenyl-functionalized porous silicon and (b) 420 molecules (700 ymol) for verapamil and 390 molecules (650 ymol) of propafenone using a bis(tridecafluoro-1,1,2,2-tetrahydrooctyl) tetramethyldisiloxane initiator
4 Mechanistic Discussions
An important question to consider is why NIMS is inherently more sensitive than traditional matrix-assisted approaches such as MALDI, especially given that these experiments are performed with the same instrumentation. While very impressive, MALDI with high sensitivity (low zeptomole) has been achieved by Keller and Li [20], MALDI however is typically 50 times less sensitive than NIMS. To assess this difference in sensitivity, sample deposition was initially examined as this is a key feature that differs between NIMS and MALDI. In typical NIMS experiments the sample droplet is spotted directly onto the nanostructured surface. The unique nonadhesive surface properties of the fluorinated modifications and coatings used for NIMS not only reduce adhesion of the analyte facilitating desorption, but also the hydrophobic nature of the coating results in the formation of small aqueous droplets that concentrate the analyte on the surface. Simply put, the aqueous droplet being hydrophilic minimizes its contact area with the fluorinated coating and dries in a smaller spot concentrating the analyte. Another advantage of this technique is in its application to real biological samples and biofluids, which often contain salts and buffers which are detrimental to mass spectrometry. The process of analyte concentration on the hydrophobic fluorinated coating separates the salts to the outer edges, essentially “cleaning up” the analyte for analysis. The hydrophobic-hydrophobic interaction occurring between the fluorocarbon and the analyte serves to corral these molecules on the nanostructured surface, minimizing the number of analyte molecules in a given area necessary to produce the analyte signal. In many cases it is possible to adsorb analyte onto the fluorous surface directly from the sample droplet to minimize the effects of interferences within the sample (e.g., salts, proteins). This is again thought to be a result of the high surface energy at the fluorous-aqueous boundary that drives adsorption of molecules with amphipathic characteristics to the interface. The concentration effect can easily improve the detection sensitivity by a factor of 10–100. This can enable a signal to be generated from a small amount of material that is quickly consumed with a few laser shots.
Another distinguishing feature between nanostructure-based desorption/ionization and MALDI is that MALDI incorporates analytes into the matrix crystals which can affect its sensitivity, as does the ionization of the matrix materials, causing analyte signal suppression. Thus in MALDI, the spatial limitation of analytes exists both laterally across the surface as well as being dependent on the matrix crystal thickness/depth (microns to millimeters in size). The resulting laser-induced ablation following each laser shot introduces new crystal surfaces from which a signal can be produced. The crystal thickness allows for a continuous signal in MALDI, yet it also introduces a dilution effect of the analyte in the matrix crystal. This dilution effect, while effective in providing a signal that continues over many laser pulses, is ultimately detrimental to achieving the highest level of sensitivity.
The length of signal duration is also quite different between nanostructure-based laser desorption/ionization and MALDI. Typically NIMS generates a signal for a significantly shorter number of laser pulses (3–50) whereas MALDI can generate a signal for hundreds if not thousands of laser shots before signal depletion occurs. The shorter signal duration characteristic of the nanostructured surfaces is likely due to the very different nature of the matrix-free nanostructure versus matrix-induced events that can occur by using MALDI. Since NIMS [14] are surface-induced phenomena, the generation of a signal is largely a 2D surface phenomenon versus 3D matrix crystals that depends on the nanosecond duration of the thermal and surface-restructuring events. Having a signal from a larger packet of ions in fewer laser shots provides a higher signal/noise ratio (S/N) since it contains a fixed amount of noise. When data is averaged over a larger number of spectra, the S/N only increases in proportion to the square root of the number of shots taken and the relatively low surface concentration in NIMS is quickly depleted. Therefore, averaging spectra from multiple laser shots ultimately results in a lower S/N than getting a larger burst of ions detected. This is analogous to LC-MS where increasing chromatographic resolution with techniques like UPLC or smaller ID columns like nano and capillary LC boosts sensitivity. Finally, an additional difference observed is in the laser energy used in NIMS (~10 mJ/cm2) which is significantly lower than that used for MALDI (~40 mJ/cm2 or higher). These lower energies can presumably reduce extraneous signal that can occur as a result of fragmentation of analyte, thereby minimizing the accumulation of background noise and improving the S/N.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree