Alternatively, gene expression can be assessed with a quantitative PCR (qPCR) based approach, in which the cDNA obtained from a sample is amplified using gene-specific primers and labeled probes. This method requires creation of specific primers and probes for each gene of interest, and logistically this limits the number of genes that can be analyzed. Oncotype DX is an example of a qPCR-based test which can be used to assess recurrence risk for women with ER-positive, lymph node-negative breast cancer. The Oncotype DX assay will be discussed further later in this chapter (Kim et al. 2009).
2.2 Validation of Gene Expression Profiling
Prior to clinical application, the use of a specific biomarker or a panel of biomarkers must be thoroughly studied and validated (Simon et al. 2009). Teutsch et al. outline three key components to evaluation of a genetic test: analytic validity, clinical validity, and (Henry and Hayes 2012; Teutsch et al. 2009). The first component, analytic validity, is defined as the ability of a test to accurately and reliably measure the genotype of interest. The analytic validity includes assessment of the sensitivity, specificity, precision (reproducibility), and assay robustness (resistance to small changes in assay parameters). The second component, clinical validity, is defined as the ability of a test to accurately and reliably predict the clinical event. This includes determination of the positive and negative predictive values of the test. Clinical validity requires analysis of an independent cohort to validate the original findings. This is often performed with an initial analysis of a “test set” and confirmation with an independent “validation set”, which may be an independent cohort of patients at the same institution or, alternatively, patients at another institution. The third component in evaluating a new biomarker is clinical utility, which is defined as evidence of improved measurable clinical outcome, and added value to patient management decision-making compared with current management. In other words, does the use of the new biomarker provide additional information that changes patient management? Also, the extent of the effect of the biomarker is considered. For example, does the biomarker status correspond to a small percentage change in outcome or does it confer a large, several-fold difference? Is this difference enough for clinicians to change management?
Microarray data analysis enables us to monitor the expression level of genes and changes in the expression patterns with respect to pathologic conditions at a genome scale. There are two approaches to analyze the data—supervised or unsupervised analysis. In supervised analysis, distinct groups of genes or samples (i.e. patient vs. normal) are identified and differences in expression profiles between the groups are evaluated. On the other hand, in unsupervised analysis, sets of genes or samples with similar expression profiles are grouped, and their common clinical and physiological and/or biological features are identified. If there are pre-existing clusters (patients and normal or different known tumor types), supervised analysis is more appropriate. However, unsupervised analysis is a powerful method for identifying new clusters (uncharacterized pathways of dysregulated gene expression, new tumor subtypes etc.) Clustering analysis generates distinct groups of genes or samples based on their similarity of expression profiles and may be hierarchical or non-hierarchical. In hierarchical clustering, the relationships among objects within and between groups are specified and represented as dendrograms (Hastie et al. 2009). In this way, samples with similar expression patterns are grouped together within branches of a sample dendrogram, and in like manner, genes with consistent expression patterns within sample groups cluster in gene dendrogram branches. These results indicate cellular and molecular features differentiating groups of samples, which may be important for diagnosis or prognosis, and furthermore identify key transcriptional and signaling pathways that may be targeted by new or existing therapies. One pitfall with microarray data analysis is the issue of multiple hypothesis testing. By definition, microarray experiments simultaneously test the expression of thousands of genes, often assessing gene expression using a much lower number of individual patient samples. This creates a statistical problem known as multiple hypothesis testing. Data analysis solutions to this problem exist, but extreme caution should be used in interpreting the results of a microarray study with relatively low numbers of patient samples (<25), thousands of gene probes and a single patient/tumor data set. The false discovery rate (FDR), which is a modified p value used to adjust for multiple comparisons, is often reported when groups of patient samples are compared for gene expression using microarrays. In general, FDR values <0.05 are acceptable for statistical significance in microarray studies, and most current studies employ additional methods to reduce the false positive rate (Benjamini and Hochberg 1995; Storey and Tibshirani 2003). In addition, a number of more advanced statistical methods are currently available for data analysis, many of which evaluate biologically meaningful gene sets or pathways (Hastie et al. 2009; Tseng et al. 2012).
In summary, prior to implementation of a new biomarker or gene signature into clinical practice, it must be evaluated for analytic validity, clinical validity, and clinical utility. Large validation studies are required. The highest level of evidence for a new genetic signature would be a prospective clinical study that is designed with assessment of the biomarker as the primary objective of the trial (Henry and Hayes 2012). This would minimize bias in subject selection and standardize sample handling and assay conditions. Gene expression profiling has been perhaps most extensively studied in breast cancer. We will next review the development and implementation of personalized medicine in breast cancer on the basis of gene expression.
3 Defining Patient Subgroups in Breast Cancer on the Basis of Gene Expression
3.1 Classical Studies of Global Gene Expression in Breast Cancer
In 2000, a seminal paper by Perou et al. characterized the gene expression patterns for 42 women with breast cancer (Perou et al. 2000). Most of the specimens analyzed were breast cancer, but a few samples of normal breast tissue were also examined. Some patients had multiple specimens studied, such as the primary tumor and a lymph node metastasis, and in some cases samples were obtained before and after chemotherapy administration. Microarrays were performed, with over 8,000 genes analyzed. Using a hierarchical clustering method, genes were grouped based on the similarity of their patterns of expression. The authors found that there was significant variation in gene expression patterns among the tumor specimens. Interestingly, samples from the same patient, such as from a primary tumor and a lymph node, or before and after chemotherapy, were more similar to each other than to any other sample, in terms of their gene expression pattern. The “intrinsic” gene subset is a subset of 496 genes which showed greater variation between unrelated samples than was seen between samples from the same patient. This subset of genes includes specific clusters of genes, such as the luminal cluster, HER2 (Erb-B2) cluster, proliferation cluster, and basal cluster. The “molecular portraits” of gene expression examined in this study led to identification of the intrinsic subtypes of breast cancer. In the first publication, the subtypes identified were ER+/luminal-like, basal-like, Erb-B2+, and normal breast (Perou et al. 2000). Further investigation led to the finding that the luminal subtype could potentially be divided into two or three subgroups, termed luminal A, luminal B, and luminal C, each with a unique gene expression pattern (Sorlie et al. 2001). Alternatively, two luminal subgroups, luminal A and luminal B, could be described. Importantly, the clinical outcome of patients was evaluated for each of the subtypes, including luminal A, luminal B, luminal C, normal breast-like, Erb-B2+, and basal-like. Significant differences were seen in both relapse-free survival and overall survival, with basal-like and Erb-B2+ subtypes having the worst outcome and luminal A subtype showing the best outcome (Fig. 2). In a subsequent analysis of 115 breast cancers, the subtypes were further refined (Sorlie et al. 2003), to include the luminal A, luminal B, basal, Erb-B2+, and normal breast-like subtypes. The cluster dendrogram for tumors, when divided into these five subtypes (luminal A, luminal B, normal breast-like, Erb-B2+, and basal-like), can be seen in Fig. 3. Also, the full cluster diagram and gene expression patterns can be seen in Fig. 4. Subtype characterization was confirmed by cluster analyses performed on independent data sets of cohorts of patients from other institutions. The clinical outcomes of different subtypes were assessed and revealed differences in overall survival and time to distant metastases. One other interesting finding from this work was that BRCA1 mutation was strongly associated with predisposition to the basal tumor phenotype.
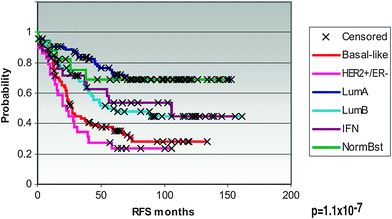
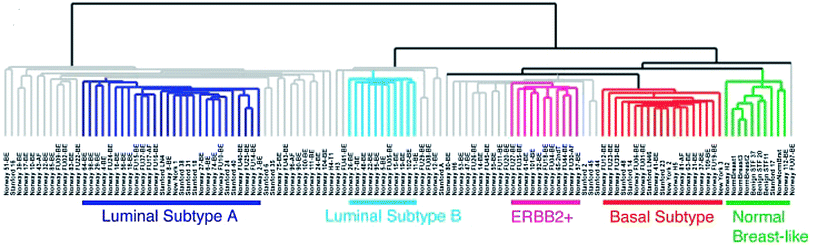
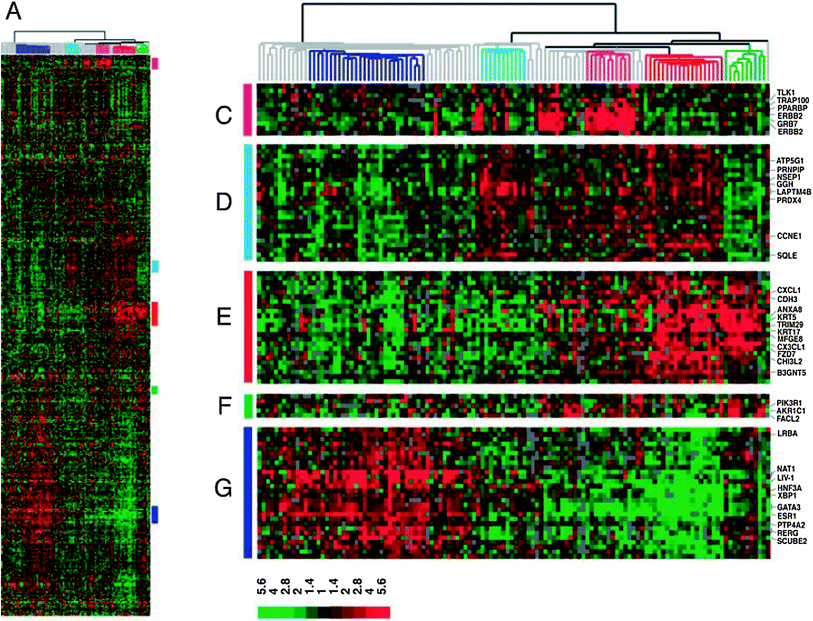

Fig. 2
Clinical outcome for patients based on tumor subtype. Kaplan-Meier survival curves demonstrate the relapse-free survival (RFS) for breast cancer patients, as classified by tumor subtype. Reprinted with permission from Hu et al. (2006)

Fig. 3
Cluster dendrogram demonstrating tumor subtypes. This dendogram shows the clustering of the tumors into five tumor subgroups. Branches for tumors with low correlation to any tumor subtype are shown in gray. Reprinted with permission from Sorlie et al. (2003)

Fig. 4
Hierarchical clustering of breast tumors using the intrinsic gene set, with full cluster (left) and specific gene clusters shown in more detail (right), including the Erb-B2+ (C), luminal B (D), basal (E), normal breast-like (F), and luminal A (G) clusters. Reprinted with permission from Sorlie et al. (2003)
More recently, a new breast cancer intrinsic gene list containing 1300 genes was evaluated and validated in independent data sets (Hu et al. 2006). This analysis stratified patients into five subtypes, luminal A, luminal B, basal-like, HER2+, and normal breast-like, using a 105 tumor training set and validation set of 311 tumor samples (compiled from three independent studies). Clinical outcomes for the subtypes were significantly different in terms of relapse-free and overall survival. Multivariate analysis demonstrated that tumor subtype was prognostic of relapse-free, disease-specific, and overall survival, independent of standard clinical factors such as tumor size, lymph node status, and tumor grade.
3.2 Implications of Breast Cancer Subtype for Systemic Therapy Selection
Determination of breast tumor subtype is clinically valuable, as the subtype (often clinically defined by immunohistochemical profile) frequently guides decisions for systemic therapy. For example, women with luminal A or luminal B (ER positive) breast cancer, are known to derive benefit from adjuvant hormone therapy (Davies et al. 2011), and therefore most will receive anti-estrogen therapy in the form of tamoxifen or an aromatase inhibitor. Patients with the Erb-B2+(HER2+) subtype will often receive Herceptin (trastuzumab), which is a monoclonal antibody against the HER2/neu receptor. Women with HER2-positive breast cancer who are either lymph node-positive or high-risk node negative have been found to benefit from Herceptin therapy (Smith et al. 2007; Perez et al. 2011) and many current studies are exploring combinations of anti-HER2 targeted therapies for these patients (Gianni et al. 2012). The basal subtype or the overlapping subtype known as “triple negative breast cancer” continues to receive significant research attention due to its relative poor prognosis and lack of targeted therapeutic strategies. Information provided by the intrinsic subtypes (using an immunohistochemical approach) has been adopted by 2011 St. Gallen Consensus Conference (Goldhirsch et al. 2011) (Table 1).
Table 1
2011 St Gallen consensus recommendations of systemic treatment
IHC Subtype | Definition | Type of adjuvant therapy |
|---|---|---|
Luminal A | HR+/HER2-/Ki67 low | Endocrine therapy alonea |
Luminal B | HR+/HER2−/Ki67 high | Endocrine therapy ± cytotoxic therapy |
HER2-positive | HR-/HER2+ | Cytotoxics + anti-HER2 therapy |
Triple-negative | HR−/HER2− | Cytotoxics |
4 Gene Signatures and Clinical Decision Making in Breast Cancer
Fortunately, the majority of women diagnosed with breast cancer do not have metastatic disease at the time of diagnosis, and are in a clinically curable situation. However, after surgery, all women have some degree of risk of relapse, at local, regional, and distant sites. The magnitude of the risk of relapse can be quite different depending on the individual woman, and can be assessed using clinical, pathologic, and treatment-related variables. Over the past decade, gene signatures for women with breast cancer have provided significant additional prognostic information. The greatest clinical impact of these assays has been risk stratification in relatively low risk patient populations (ER positive and node negative). Specifically, Mammaprint is a commercially available gene expression assay that has been used to predict recurrence in patients with node negative cancers. Oncotype DX is a commercially available, clinically validated gene expression assay that is used to guide recommendations for the use of systemic chemotherapy in addition to anti-estrogen therapy for patients with ER positive, node negative disease. It is important to note that clinical use of any gene signature should be considered only for independently validated assays performed on large datasets, and care should be taken to apply use of the signature only to patients for whom the assay has been validated. Currently, clinical use of Oncotype DX in node positive and/or ER negative disease is considered experimental.
4.1 Mammaprint
One of the earliest gene signatures for breast cancer was the Mammaprint, or the Amsterdam 70-gene prognostic signature. This signature was initially characterized in a cohort of 98 primary breast cancer patients, which included 34 women that developed distant metastatic disease within 5 years and 44 women who did not (van’t Veer et al. 2002). There were also 20 patients with BRCA1 or BRCA2 germline mutations included in the analysis. For the sporadic cases, all women were <55 years old, had a tumor size <5 cm, and were lymph node negative. RNA was isolated from frozen tumor samples and supervised analysis of the microarray data identified a set of 70 genes that allowed discrimination between patients with good and poor prognosis, with an accuracy of 83 %. The Mammaprint allows a binary classification, either a good prognosis or poor prognosis signature (Fig. 5). Women in the poor prognostic group based on this signature have a significantly increased risk of developing distant metastatic disease within 5 years (odds ratio, OR = 28). On multivariate analysis including classical prognostic factors, the Mammaprint signature was an independent predictor of outcome.
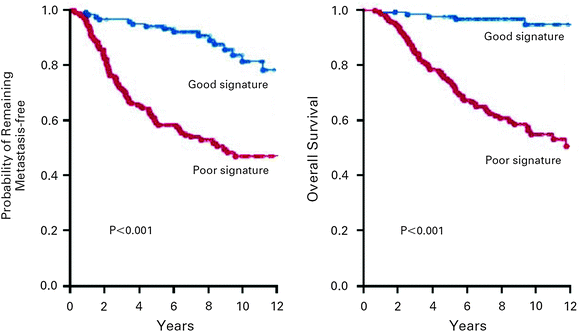

Fig. 5
Patient outcome based on Mammaprint signature. The metastasis-free survival (left panel) and overall survival (right panel) of breast cancer patients with good prognosis signature and poor prognosis signature are shown. Reprinted with permission from van de Vijver et al. (2002)
Further validation of the Mammaprint gene signature was performed in a cohort of 295 patients (van de Vijver et al. 2002). All patients were less than 53 years old, had a primary tumor size of less than 5 cm, and in this cohort 151 were lymph node negative, while 144 were lymph node positive. Overall survival at 10 years was 95 % for those with good prognosis signature and was 55 % for those with poor prognosis signature. Probability of remaining free of distant metastases was 85 % for those with good prognosis signature and 51 % for those with poor prognosis signature. However, it should be noted that 61 patients in this study were also members of the original cohort used to develop the signature. Independent validation studies were subsequently conducted, including an analysis of 302 patients from five European centers (Buyse et al. 2006). Patients included in this analysis were <61 years old, lymph node negative, tumor size <5 cm, and did not receive adjuvant systemic therapy. Median follow-up was 13.6 years. The 70-gene prognostic signature remained an independent prognostic factor for development of distant metastases and overall survival, with unadjusted hazard ratios of 2.32 and 2.79, respectively.
There is also interest in assessing risk of distant metastatic disease in older women, who may not tolerate chemotherapy well. Identification of older women that have a low risk of distant metastatic disease might allow avoidance of the considerable toxicity associated with chemotherapy in this group of women. The Mammaprint gene signature has also been evaluated for women between the ages of 55 and 70 (Mook et al. 2010). In this analysis, frozen tumor specimens from 148 women, aged 55−70 years old with tumor size <5 cm and negative lymph nodes, were analyzed and assigned either good or poor prognosis based on their 70-gene signature. The 70-gene prognosis signature was prognostic of breast cancer-specific survival (P = 0.036). Distant metastasis-free survival at 5 years was 93 % for patients with a good prognosis signature and 72 % for those with poor prognosis signature, but this difference was not statistically different in this cohort (P = 0.07).
Currently, there are several ongoing clinical trials designed to further characterize the utility of Mammaprint. MINDACT (Microarray In Node-negative and 1−3 positive lymph node Disease may Avoid ChemoTherapy; EORTC 10041), is a Phase III prospective randomized study comparing Mammaprint with clinical-pathological assessment (Adjuvant! Online) in selecting patients with 0−3 positive lymph nodes for adjuvant chemotherapy. In the trial, women with discordant prognostic assessment on Mammaprint and Adjuvant! Online will be randomized for the decision of adjuvant chemotherapy based on either Mammaprint or Adjuvant! Online risk status. All women with high risk scores on both Mammaprint and Adjuvant! Online will receive chemotherapy and all women with low risk scores on both will not receive chemotherapy. Accrual of 6,600 patients has been achieved, with results currently pending.
Other ongoing trials include PROMIS, which is a prospective registry study to assess the impact of Mammaprint on systemic therapy decision making for patients with an intermediate Oncotype DX score. NBRST, is a prospective registry study designed to measure outcomes based on molecular subgroups, determined by Mammaprint and other profiles, for patients undergoing neoadjuvant chemotherapy or endocrine therapy. Similarly, MINT I, is a study designed to test the ability of Mammaprint (in combination with other factors) to predict response to neoadjuvant chemotherapy. Of note, initial studies using Mammaprint were conducted on frozen tissue, but currently the assay can be performed on fresh, frozen, or formalin-fixed paraffin-embedded specimens.
4.2 Oncotype DX
The Oncotype DX is a real-time quantitative reverse-transcriptase-polymerase-chain-reaction (RT-PCR) assay of 21 prospectively selected genes designed for use in fixed, paraffin-embedded tumor specimens. Paik and colleagues first developed a real-time RT-PCR assay to quantify gene expression of 250 candidate genes and subsequently analyzed the relation between breast cancer recurrence and gene expression in a preliminary inquiry of 447 patients (Paik et al. 2004). From this analysis, they selected a panel of 16 cancer-related genes and five reference genes. The cancer-related genes included those involved in proliferation and invasion, among others. An algorithm was designed to calculate a Recurrence Score (RS) based on the levels of expression of these genes. The RS ranged from 0 to 100, with higher scores reflecting greater likelihood of distant recurrence. Patients were divided into three risk categories based on their RS: low-risk (RS < 18), intermediate-risk (RS 18−30), and high-risk (RS > 30). Paik et al. demonstrated the ability of the Oncotype DX assay to predict the likelihood of distant recurrence for women with ER-positive, lymph node-negative breast cancer (Paik et al. 2004). In this analysis, tumor samples from 668 women treated with tamoxifen on NSABP B-14 were evaluated. NSABP B-14 was a clinical trial of ER-positive, node-negative breast cancer, in which women were randomized to tamoxifen versus placebo. The rate of distant recurrence at 10 years was 7 % for those with low-risk RS, 14 % for intermediate-risk RS, and 31 % for high-risk RS (Fig. 6). Recurrence score was found to be an independent prognostic factor on multivariate analysis. RS was also predictive of overall survival. The risk of distant recurrence can be predicted using the RS as a continuous function (Fig. 7).
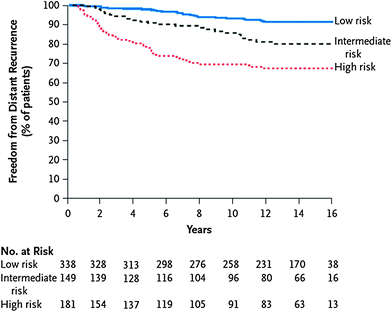
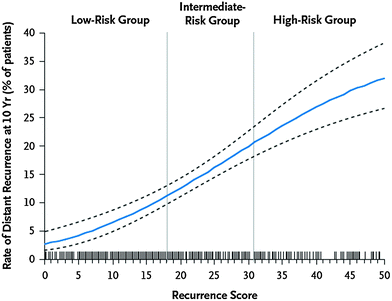

Fig. 6
Stratification of patient outcome based on Oncotype DX risk group. The freedom from distant recurrence is shown for patients in the low, intermediate, and high risk groups. Reprinted with permission from Paik et al. (2004)

Fig. 7
Rate of distant metastases as a function of Recurrence Score, determined by Oncotype DX analysis. The risk of distant metastasis at 10 years can be estimated for any given recurrence score, using the continuous function shown here. Reprinted with permission from Paik et al. (2004)
Determination of the RS from the Oncotype DX assay was also found to predict the magnitude of benefit from chemotherapy. An analysis was performed of tumor samples from the NSABP B-20 trial, in which women with ER-positive, node negative breast cancer were randomized to tamoxifen with or without chemotherapy. RT-PCR was successful in the majority (97 %) of blocks that had sufficient remaining specimen and were therefore included in this analysis (n = 651). In NSABP B-20, there was a benefit with the addition of chemotherapy, in terms of local, regional, and distant recurrence. In the current study, the magnitude of benefit from chemotherapy was greatest among patients with high risk RS, and patients in intermediate or low risk groups demonstrated no significant benefit from chemotherapy (Paik et al. 2006). In another study, the Oncotype DX-derived RS was found to be predictive of pathologic complete response (pCR) in patients receiving neoadjuvant chemotherapy (Gianni et al. 2005). Patients with higher RS had a greater probability of having a pCR after completion of neoadjuvant chemotherapy. Based on the findings above, the randomized Phase III TAILORx trial for women with ER-positive, node-negative breast cancer was designed. Women enrolled on this trial with an Oncotype DX RS <11 receive hormone therapy only, and those with a RS >25 receive chemotherapy followed by hormone therapy. Women with RS of 11−25 are randomized to either hormone therapy alone or in combination with chemotherapy. Enrollment has been completed and results are currently pending.
Two preliminary studies have reported on the utility of the Oncotype DX assay in lymph-node positive patients (Dowsett et al. 2010; Albain et al. 2010). Dowsett et al. evaluated the Oncotype DX assay on specimens from the ATAC (Arimidex, Tamoxifen, Alone or in Combination) trial (Dowsett et al. 2010). The ATAC trial compared the efficacy of arimidex, tamoxifen, or both for post-menopausal women. In this analysis, Oncotype DX assay was performed for samples of patients on the single agent arms (arimidex only or tamoxifen only; n = 1,372). Prognostic value of Oncotype-based RS was seen both in patients treated with tamoxifen and confirmed for patients treated with arimidex. In addition, RS was predictive of distant recurrence for lymph node negative (n = 872) and also for lymph node positive patients (n = 306). Similarly, Albain et al. investigated the utility of the Oncotype DX assay for post-menopausal women with node-positive, ER-positive breast cancer (Albain et al. 2010). In this retrospective analysis, 367 samples from the Phase III SWOG-8814 trial, in which women were randomized to tamoxifen with or without the addition of chemotherapy, were analyzed. Patients with high-risk RS showed a significant benefit in disease-free survival with the addition of chemotherapy, whereas those with intermediate- or low-risk RS did not. An ongoing Phase III trial, SWOG RxPONDER (S1007), has been initiated to prospectively evaluate the benefit of chemotherapy for women with 1−3 positive lymph nodes, ER-positive, HER2-negative breast cancer with RS of 25 or less. Women meeting eligibility criteria will be randomized to receive endocrine therapy with or without the addition of chemotherapy. The primary endpoint is disease-free survival and enrollment is ongoing.
Oncotype DX assessment is incorporated in the ASCO recommendations for use of tumor markers in breast cancer and in NCCN guidelines to predict the risk of recurrence for women with ER-positive, HER2-negative, node-negative breast cancer treated with tamoxifen. The NCCN panel considers Oncotype DX an option, to be taken into consideration only in the context of other elements of patient risk stratification. Other applications at this time are investigational and require further validation prior to inclusion in clinical decision-making.
Recently there has been interest in molecular characterization of ductal carcinoma in situ, DCIS. DCIS is a non-invasive form of breast cancer that has the potential to develop into invasive disease over time. Adjuvant radiation therapy has been shown to decrease the risk of both non-invasive and invasive recurrence after partial mastectomy for DCIS. There is ongoing interest in identifying women with DCIS with a low risk of recurrence after surgery who would likely derive a small absolute benefit from adjuvant therapy. However, the use of clinical and pathologic factors to identify women with low risk DCIS is not entirely straightforward. Even patients with low or intermediate grade DCIS and negative margins have a measurable risk of recurrence. Solin et al. reported DCIS risk stratification utilizing 12 of the 21 genes in the Oncotype DX assay (Solin et al. 2012). In data presented at the San Antonio Breast Cancer Symposium in 2011, the Oncotype DX assay has been proposed as a means to stratify risk of recurrence in patients with DCIS who have undergone partial mastectomy alone. For this analysis, the Oncotype DX was performed for 327 women (approximately 50 % of patients) enrolled on the ECOG E-5194 study. Women in this study had relatively low risk DCIS, with either low or intermediate grade DCIS ≤ 2.5 cm or high grade DCIS ≤ 1 cm. The “DCIS Score” was determined and women were grouped into three categories: high, intermediate, and low. They found that the DCIS Score obtained from the Oncotype DX assay was a significant independent predictor of ipsilateral breast events (either invasive or DCIS) on multivariate analysis. It was proposed that the DCIS Score could be used to estimate a woman’s risk of recurrence after partial mastectomy for DCIS and potentially guide individual recommendations for adjuvant therapy. However, even the low risk group had a 12 % risk of any breast event (either invasive or non-invasive recurrence). Also, further validation is necessary before this is applied to routine clinical practice.
4.3 Other Novel Gene Signature Assays
Other gene signatures are being developed, including the Breast Cancer Index (BCI), MapQuant DX Genomic Grade Index (GGI), among others (Prat et al. 2011). The Breast Cancer Index (BCI) is an assay for women with ER-positive, node-negative breast cancer, which may assist in prediction of the risk of distant recurrence. Two prognostic quantitative real-time PCR assays, the HOXB13:IL17BR two-gene ratio and the molecular grade index (MGI), are incorporated in the BCI. HOXB13 is an antiapoptotic gene associated with increased risk of recurrence and the IL-17 receptor B is associated with decreased risk of recurrence. The combination of the HOXB13:IL17BR and MGI were found to be prognostic of outcome (Ma et al. 2008). MapQuant DX, also known as Genomic Grade Index or GGI, is a microarray based assay that defines two molecular subgroups that have distinct clinical outcomes (Loi et al. 2007). Early comparisons of the various gene signatures show high rates of concordance in predicting outcome (Fan et al. 2006).
4.4 Gene Expression and Radiation Therapy
The primary purpose and utility of the previously discussed gene signatures was to stratify patients based on risk of distant recurrence. There are several questions regarding gene expression in breast cancer that are more relevant to radiation therapy, and we will review the existing data for each of these questions here. Haffty and Buchholz (2010) have recently written an excellent editorial on recent publications concerning gene expression and local recurrence in breast cancer, and interested readers are encouraged to review their findings (Haffty and Buchholz 2010). In brief, the authors define the following questions regarding gene expression and radiation therapy: (1) Is there a group of women at sufficiently low risk of local recurrence after lumpectomy alone that can be spared radiation therapy? (2) Alternatively, can gene expression signatures be used to identify a group of women at sufficiently high risk of local or regional recurrence after breast conservation (BCT)? Would these patients be better served with dose escalation or the use of radiosensitizers? (3) As breast conservation treatments evolve to include accelerated regimens, hypofractionation and partial breast volumes, can the results of gene expression assist in selection of patients for altered radiotherapy regimens? (4) With respect to postmastectomy irradiation, are our current guidelines oversimplified with respect to tumor biology? Can gene expression profiling be used to identify patients at risk for local regional recurrence after mastectomy?
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree