92 % but with high FP level ( 50 per patient). Regions of interest (ROI) for lesion candidates are generated in this step and function as input for the second tier. In the second tier we generate
50 per patient). Regions of interest (ROI) for lesion candidates are generated in this step and function as input for the second tier. In the second tier we generate 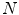 2D views, via scale, random translations, and rotations with respect to each ROI centroid coordinates. These random views are used to train a deep Convolutional Neural Network (CNN) classifier. In testing, the CNN is employed to assign individual probabilities for a new set of
2D views, via scale, random translations, and rotations with respect to each ROI centroid coordinates. These random views are used to train a deep Convolutional Neural Network (CNN) classifier. In testing, the CNN is employed to assign individual probabilities for a new set of 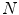 random views that are averaged at each ROI to compute a final per-candidate classification probability. This second tier behaves as a highly selective process to reject difficult false positives while preserving high sensitivities. We validate the approach on CT images of 59 patients (49 with sclerotic metastases and 10 normal controls). The proposed method reduces the number of FP/vol. from 4 to 1.2, 7 to 3, and 12 to 9.5 when comparing a sensitivity rates of 60, 70, and 80 % respectively in testing. The Area-Under-the-Curve (AUC) is 0.834. The results show marked improvement upon previous work.
random views that are averaged at each ROI to compute a final per-candidate classification probability. This second tier behaves as a highly selective process to reject difficult false positives while preserving high sensitivities. We validate the approach on CT images of 59 patients (49 with sclerotic metastases and 10 normal controls). The proposed method reduces the number of FP/vol. from 4 to 1.2, 7 to 3, and 12 to 9.5 when comparing a sensitivity rates of 60, 70, and 80 % respectively in testing. The Area-Under-the-Curve (AUC) is 0.834. The results show marked improvement upon previous work.
1 Introduction
2 Methods
2.1 Sclerotic Metastases Candidate Detection
2.2 CNN Training on 2D Image Patches
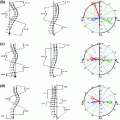
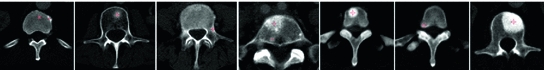
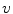 in axial space. Furthermore, each translated ROI is rotated around its center
in axial space. Furthermore, each translated ROI is rotated around its center 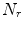 times by a random angle
times by a random angle ![$$\alpha = [0^{\circ },\ldots ,360^{\circ }]$$](/wp-content/uploads/2016/10/A331518_1_En_1_Chapter_IEq7.gif) . These translations and rotations for each ROI are computed
. These translations and rotations for each ROI are computed 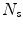 times at different physical scales
times at different physical scales 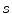 (the edge length of each ROI), but with fixed numbers of pixels. This procedure results in
(the edge length of each ROI), but with fixed numbers of pixels. This procedure results in 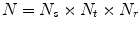 random observation of each ROI—an approach similar to [21]. Note that 2.5–5 mm thick-sliced CT volumes are used for this study. Due to this relative large slice thickness, our spatial transformations are all drawn from within the axial plane. This is in contrast to other approaches that use CNNs which sample also sagittal and/or coronal planes [13, 14]. Following this procedure, both the training and test data can be easily expanded to better scale to this type of neural net application. A CNN’s predictions on these
random observation of each ROI—an approach similar to [21]. Note that 2.5–5 mm thick-sliced CT volumes are used for this study. Due to this relative large slice thickness, our spatial transformations are all drawn from within the axial plane. This is in contrast to other approaches that use CNNs which sample also sagittal and/or coronal planes [13, 14]. Following this procedure, both the training and test data can be easily expanded to better scale to this type of neural net application. A CNN’s predictions on these 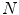 random observations
random observations 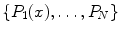 can then be simply averaged at each ROI to compute a per-candidate probability:
can then be simply averaged at each ROI to compute a per-candidate probability: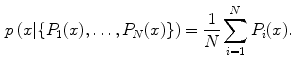
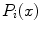 is the CNN’s classification probability computed one individual image patch. In theory, more sophisticated fusion rules can be explored but we find that simple averaging works well. This proposed random resampling is an approach to effectively and efficiently increase the amount of available training data. In computer vision, translational shifting and mirroring of 2D image patches is often used for this purpose [11]. By averaging the
is the CNN’s classification probability computed one individual image patch. In theory, more sophisticated fusion rules can be explored but we find that simple averaging works well. This proposed random resampling is an approach to effectively and efficiently increase the amount of available training data. In computer vision, translational shifting and mirroring of 2D image patches is often used for this purpose [11]. By averaging the 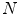 predictions on random 2D views as in Eq. 1, the robustness and stability of CNN can be further increased as shown in Sect. 3.
predictions on random 2D views as in Eq. 1, the robustness and stability of CNN can be further increased as shown in Sect. 3.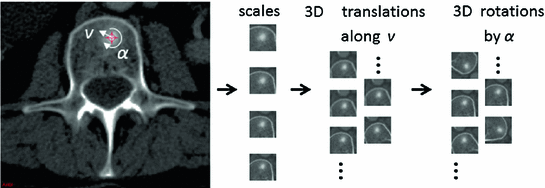
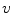 ) and rotations (by a random angle
) and rotations (by a random angle 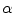 ) in the axial plane
) in the axial planeStay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree