and Ron M. A. Heeren1
(1)
Maastricht MultiModal Molecular Imaging (M4I) Institute, Maastricht University, Universiteitssingel 50, 6229, ER, Maastricht, The Netherlands
(2)
Physical Electronics, Inc., Chanhassen, Minnesota 55317, USA
Abstract
Unambiguous identification of detected species is essential in complex biomedical samples. To date, there are not many mass spectrometry imaging techniques that can provide both high spatial resolution and identification capabilities. A new and patented imaging tandem mass spectrometer, exploiting the unique characteristics of the nanoTOF II (Physical Electronics, USA) TOF-SIMS TRIFT instrument, was developed to address this.
Tandem mass spectrometry is based on the selection of precursor ions from the full secondary ion spectrum (MS1), followed by energetic activation and fragmentation, and collection of the fragment ions to obtain a tandem MS spectrum (MS2). The PHI NanoTOF II mass spectrometer is equipped with a high-energy collision induced dissociation (CID) fragmentation cell as well as a second time-of-flight analyzer developed for simultaneous ToF-SIMS and tandem MS imaging experiments.
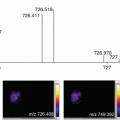
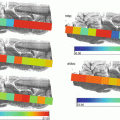
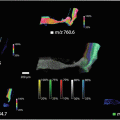
We describe here the results of a ToF-SIMS imaging experiment on a thin tissue section of an infected zebrafish as a model organism for tuberculosis. The focus is on the obtained ion distribution plot of a fatty acid as well as its identification by tandem mass spectrometry.
Key words
SIMSTandem MSImaging mass spectrometryLipidsFatty acids1 Introduction
Time-of-flight secondary ion mass spectrometry (ToF-SIMS) analysis depends on a high energy focused ion beam that functions as a primary ion source ejecting analytes only from the top atomic monolayers of a sample surface, referred to as secondary ions. The secondary ions are separated in a time-of-flight mass analyzer based on their mass-to-charge ratio (m/z) before they hit the detector [1–3].
Traditionally, ToF-SIMS has been applied in the domain of surface physics and semiconductor industry, but over the last decade it has redirected toward biomedical applications as well. Especially in mass spectrometry imaging -based biological tissue analysis, one has to contend with a high level of complexity concerning the molecules that are found on the sample surface. In many cases, it is predictable and generally accepted which compounds are detected in specific samples . Although the mass resolution and mass accuracy in ToF analyzers have improved over the years, particularly for higher masses this is not sufficient for unambiguous peak identification.
Tandem mass spectrometry is considered the benchmark technique to identify and elucidate structures of unknown molecular species and distinguish isomers of which there are multiplicities in biological samples. Controlled fragmentation of ions after they are formed in the source of a mass spectrometer results in a unique fragment pattern for every compound and is therefore an extremely useful tool for identification and elucidation of molecules.
Matrix-assisted laser desorption/ionization (MALDI) is often the ionization technique of choice for tandem MS imaging in biological tissues, but has several inherent limitations compared to ToF-SIMS imaging. First, in contrast to ToF-SIMS, MALDI needs a homogeneously applied chemical matrix to absorb the laser energy, to extract and ionize compounds from the sample. This makes the sample preparation time consuming and results in the perturbation of the intrinsic molecular chemistry and loss of lateral resolution . Furthermore, photon ionization of the analytes in a MALDI ion source is very destructive and usually consumes the sample, whereas in ToF-SIMS only sub nanometer thick layers are removed from the sample surface. In addition, the typical spatial resolution of MALDI imaging is above 30 μm, ToF-SIMS imaging can reach submicron spatial resolution. Lastly, current instrument designs do not allow simultaneous collection of MS1 and MS2 imaging data sets.
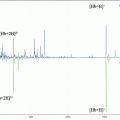
Recently, a novel and patented PHI NanoTOF II TRIFT mass spectrometer was developed, equipped with a second time-of-flight analyzer designed for tandem MS imaging [4–6]. The ion optics of the first mass analyser allow selecting a precursor from the stream of generated ions to be analyzed in the tandem time-of-flight mass analyzer simultaneously. This makes the combination of surface screening with general MS imaging with targeted identification with tandem MS imaging possible (Fig. 1).


Fig. 1
(a) Schematic illustration of the parallel imaging MS/MS spectrometer with the MS1 (TRIFT) and the MS2 (linear TOF) analysers as well as their detectors annotated. The arrows indicate the nominal trajectories of the secondary ions (in red). The precursor selection device including the activation cell is framed in (a) and enlarged in (b)
In this chapter, we show an example of a tandem TOF-SIMS imaging experiment performed on a whole-body section prepared from a diseased zebrafish. The fish was part of a study on tuberculosis and therefore infected with Mycobacterium marinum as a model for human tuberculosis infection. Without further preparation, the samples were analyzed in negative ion mode using a PHI nanoTOF II TOF-TOF mass spectrometer equipped with a liquid metal ion gun (Bi3 +), a gas collision cell, and a second ToF detector. ToF-SIMS tandem imaging MS was used to localize and identify lipid species and fatty acids as well as the host membrane phospholipids that are believed to play an important role in the bacterial inflammation cascade.
2 Materials
2.1 Chemicals
- 1.
Dietrich’s fixative (30% ethanol, 10% formalin, 2% glacial acetic acid in deionized water).
- 2.
Mixture of 5% carboxyl methyl cellulose (~90,000 Da polymer length) and 10% porcine gelatine, kept liquid at 37 °C.
- 3.
Dry ice.
- 4.
Ethanol.
- 5.
Indium tin oxide (ITO)-coated glass slides .
- 6.
Starfrost glass slides.
- 7.
Acetone (at −20 °C).
- 8.
Eosin.
- 9.
Hematoxylin.
- 10.
MilliQ water.
- 11.
95% ethanol.
- 12.
Pure ethanol (96%).
- 13.
Glycerol.
- 14.
Cover slip.
2.2 Instrumentation
- 1.
Cryotome at −35 °C.
- 2.
Mirax slide scanner (Carl Zeiss).
- 3.
PHI nanoTOF II TOF-SIMS instrument (Physical Electronics, Minnesota, U.S.A.) equipped with a 30 kV Bin q+ cluster liquid metal ion gun (LMIG), a precursor selection device, a collision cell, and a parallel linear time-of-flight tube suitable to perform tandem mass spectrometry.
2.3 Software
- 1.
The nanoTOF MS/MS instrument is controlled by SmartSoft version 2.1.0 (PHI, USA).
- 2.
TOF-DR version 1.1.0.1. (PHI, USA) was used for spectral and image analysis.
3 Methods
Zebrafish (Danio rerio) are considered a suitable model to study infectious diseases such as tuberculosis (TB) and to aid in the development of treatments [7]. Mycobacterium marinum bacteria cause tuberculosis in fish, and has a very similar disease process compared to Mycobacterium tuberculosis, which is responsible for tuberculosis infections in humans.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree