by using high-dimensional data 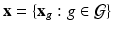 measured on a graph
measured on a graph 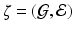 , where
, where 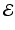 is the edge set of
is the edge set of 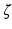 and
and 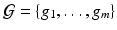 is a set of vertexes, in which m is the total number of vertexes in
is a set of vertexes, in which m is the total number of vertexes in 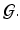 The response
The response 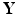 may include cognitive outcome, disease status, and the early onset of disease, among others. Standard graphs including both directed and undirected graphs have been widely used to build complex patterns [10]. Examples of graphs are linear graphs, tree graphs, triangulated graphs, and 2-dimensional (2D) (or 3-dimensional (3D)) lattices, among many others (Fig. 1). Examples of
may include cognitive outcome, disease status, and the early onset of disease, among others. Standard graphs including both directed and undirected graphs have been widely used to build complex patterns [10]. Examples of graphs are linear graphs, tree graphs, triangulated graphs, and 2-dimensional (2D) (or 3-dimensional (3D)) lattices, among many others (Fig. 1). Examples of 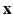 on the graph
on the graph 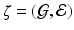 include time series and genetic data measured on linear graphs and imaging data measured on triangulated graphs (or lattices). Particularly, various structural and functional neuroimaging data are frequently measured in a 3D lattice for the understanding of brain structure and function and their association with neuropsychiatric and neurodegenerative disorders [9].
include time series and genetic data measured on linear graphs and imaging data measured on triangulated graphs (or lattices). Particularly, various structural and functional neuroimaging data are frequently measured in a 3D lattice for the understanding of brain structure and function and their association with neuropsychiatric and neurodegenerative disorders [9].
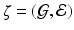 : (a) two-dimensional lattice; (b) acyclic directed graph; (c) tree; (d) undirected graph.
: (a) two-dimensional lattice; (b) acyclic directed graph; (c) tree; (d) undirected graph. on graph
on graph 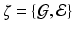 to predict
to predict 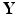 . Four major challenges arising from such development include ultra-high dimensionality, low sample size, spatially correlation, and spatial smoothness. SWPCR is developed to address these four challenges when high-dimensional data on graphs
. Four major challenges arising from such development include ultra-high dimensionality, low sample size, spatially correlation, and spatial smoothness. SWPCR is developed to address these four challenges when high-dimensional data on graphs 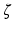 share two important features including spatial smoothness and intrinsically low dimensional structure. Compared with the existing literature, we make several major contributions as follows:
share two important features including spatial smoothness and intrinsically low dimensional structure. Compared with the existing literature, we make several major contributions as follows:
(i) SWPCR is designed to efficiently capture the two important features by using some recent advances in smoothing methods, dimensional reduction methods, and sparse methods.
(ii) SWPCR provides a powerful dimension reduction framework for integrating feature selection, smoothing, and feature extraction.
(iii) SWPCR significantly outperforms the competing methods by simulation studies and the real data analysis.
2 Spatially Weighted Principal Component Regression
2.1 Graph Data
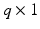 vector of discrete or continuous responses, denoted by
vector of discrete or continuous responses, denoted by 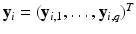 , and a
, and a 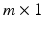 vector of high dimensional data
vector of high dimensional data 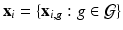 for
for 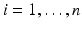 . In many cases, q is relatively small compared with n, whereas m is much larger than n. For instance, in many neuroimaging studies, it is common to use ultra-high dimensional imaging data to classify a binary class variable. In this case,
. In many cases, q is relatively small compared with n, whereas m is much larger than n. For instance, in many neuroimaging studies, it is common to use ultra-high dimensional imaging data to classify a binary class variable. In this case, 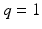 , whereas m can be several million number of features. In many applications,
, whereas m can be several million number of features. In many applications, 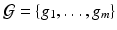 is a set of prefixed vertexes, such as voxels in 2D or 3D lattices, whereas the edge set
is a set of prefixed vertexes, such as voxels in 2D or 3D lattices, whereas the edge set 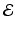 may be either prefixed or determined by
may be either prefixed or determined by 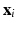 (or other data).
(or other data).2.2 SWPCR
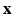 to predict a set of response variables
to predict a set of response variables 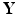 . The key stages of SWPCR can be described as follows.
. The key stages of SWPCR can be described as follows.
Stage 1. Build an importance score vector (or function)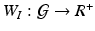 and the spatial weight matrix (or function)
and the spatial weight matrix (or function) 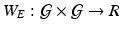 .
.
Stage 2. Build a sequence of scale vectors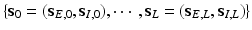 ranging from the smallest scale vector
ranging from the smallest scale vector 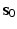 to the largest scale vector
to the largest scale vector 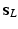 . At each scale vector
. At each scale vector 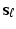 , use generalized principal component analysis (GPCA) to compute the first few principal components of an
, use generalized principal component analysis (GPCA) to compute the first few principal components of an 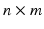 matrix
matrix 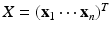 , denoted by
, denoted by 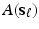 , based on
, based on 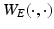 and
and 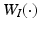 for
for 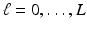 .
.
Stage 3. Select the optimal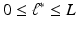 and build a prediction model (e.g., high-dimensional linear model) based on the extracted principal components
and build a prediction model (e.g., high-dimensional linear model) based on the extracted principal components 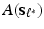 and the responses
and the responses 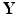 .
.
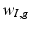 play an important feature screening role in SWPCR. Examples of
play an important feature screening role in SWPCR. Examples of 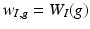 in the literature can be generated based on some statistics (e.g., Pearson correlation or distance correlation) between
in the literature can be generated based on some statistics (e.g., Pearson correlation or distance correlation) between 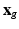 and
and 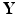 at each vertex g. For instance, let p(g) be the Pearson correlation at each vertex g and then define
at each vertex g. For instance, let p(g) be the Pearson correlation at each vertex g and then define![$$\begin{aligned} w_{I,g}=- m \; \mathrm{log} ({p(g)})/\left[ -\sum _{g \in \mathcal G}\mathrm{log} ({p(g)})\right] . \end{aligned}$$](/wp-content/uploads/2016/09/A339424_1_En_60_Chapter_Equ1.gif)
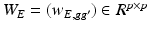 throughout the paper. The element
throughout the paper. The element 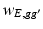 is usually calculated by using various similarity criteria, such as Gaussian similarity from Euclidean distance, local neighborhood relationship, correlation, and prior information obtained from other data [21]. In Sect. 2.3, we will discuss how to determine
is usually calculated by using various similarity criteria, such as Gaussian similarity from Euclidean distance, local neighborhood relationship, correlation, and prior information obtained from other data [21]. In Sect. 2.3, we will discuss how to determine 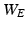 and
and 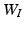 while explicitly accounting for the complex spatial structure among different vertexes.
while explicitly accounting for the complex spatial structure among different vertexes.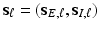 , we construct two matrices, denoted by
, we construct two matrices, denoted by  and
and 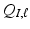 based on
based on 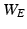 and
and 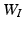 as follows:
as follows: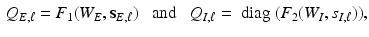
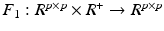 and
and 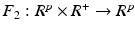 are two known functions. For instance, let
are two known functions. For instance, let 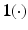 be an indicator function, we may set
be an indicator function, we may set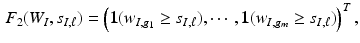
 . For instance, one may set
. For instance, one may set  as
as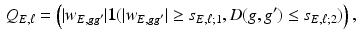
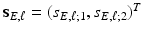 and
and 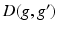 is a graph-based distance between vertexes g and
is a graph-based distance between vertexes g and 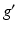 . The value of
. The value of 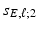 controls the number of vertexes in
controls the number of vertexes in 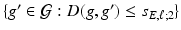 , which is a patch set at vertex g [18], whereas
, which is a patch set at vertex g [18], whereas 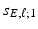 is used to shrink small
is used to shrink small 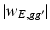 s into zero.
s into zero. and
and 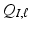 , we set
, we set 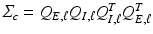 and
and 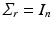 for independent subjects. Let
for independent subjects. Let 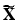 be the centered matrix of
be the centered matrix of 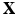 . Then we can extract K principal components through minimize the following objective function given by
. Then we can extract K principal components through minimize the following objective function given by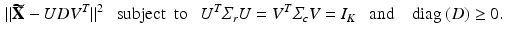
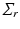 to explicitly model their correlation structure. The solution
to explicitly model their correlation structure. The solution 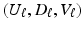 of the objective function (4) at
of the objective function (4) at 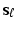 is the SVD of
is the SVD of 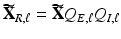 . The we can use a GPCA algorithm to simultaneously calculate all components of
. The we can use a GPCA algorithm to simultaneously calculate all components of 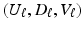 for a fixed K as follows. In practice, a simple criterion for determining K is to include all components up to some arbitrary proportion of the total variance, say 85
for a fixed K as follows. In practice, a simple criterion for determining K is to include all components up to some arbitrary proportion of the total variance, say 85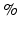 .
.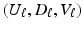 by minimizing the following objective function
by minimizing the following objective function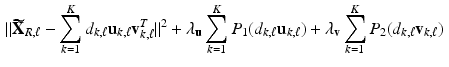
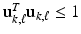 and
and 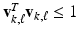 for all k, where
for all k, where 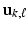 and
and 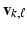 are respectively the k-th column of
are respectively the k-th column of 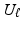 and
and 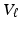 . We use adaptive Lasso penalties for
. We use adaptive Lasso penalties for 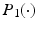 and
and 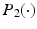 and then iteratively solve (5) [1]. For each
and then iteratively solve (5) [1]. For each 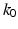 , we define
, we define 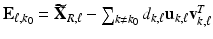 and minimize
and minimize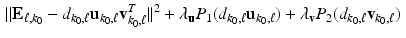
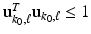 and
and 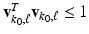 . By using the sparse method in [12], we can calculate the solution of (6), denoted by
. By using the sparse method in [12], we can calculate the solution of (6), denoted by 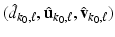 . In this way, we can sequentially compute
. In this way, we can sequentially compute 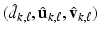 for
for 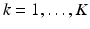 .
.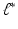 as the minimum point of the objective function (5) or (6) . let
as the minimum point of the objective function (5) or (6) . let 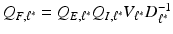 and then K principal components
and then K principal components 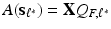 . Moreover, K is usually much smaller than
. Moreover, K is usually much smaller than  . Then, we build a regression model with
. Then, we build a regression model with 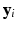 as responses and
as responses and 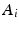 (the i-th row of
(the i-th row of 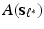 ) as covariates, denoted by
) as covariates, denoted by 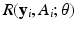 , where
, where 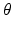 is a vector of unknown (finite-dimensional or nonparametric) parameters. Specifically, based on
is a vector of unknown (finite-dimensional or nonparametric) parameters. Specifically, based on 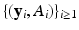 , we use an estimation method to estimate
, we use an estimation method to estimate 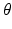 as follows:
as follows: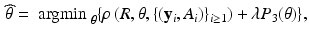
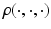 is a loss function, which depends on both the regression model and the data, and
is a loss function, which depends on both the regression model and the data, and 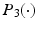 is a penalty function, such as Lasso. This leads to a prediction model
is a penalty function, such as Lasso. This leads to a prediction model 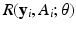 . For instance, for binary response
. For instance, for binary response 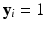 or 0, we may consider a sparse logistic model given by
or 0, we may consider a sparse logistic model given by 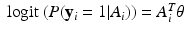 for
for 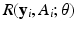 .
.Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree




