Gene expression is a process of DNA sequence reading into protein synthesis. In cases of problems in DNA repair/apoptosis mechanisms, cells accumulate genomic abnormalities and pass them through generations of cells. The accumulation of mutations causes diseases and even tumors. In addition to cancer, many other neurologic conditions have been associated with genetic mutations. Some trials are testing patients with epigenetic treatments. Epigenetic therapy must be used with caution because epigenetic processes and changes happen constantly in normal cells, giving rise to drug off-target effects. Scientists are making progress in specifically targeting abnormal cells with minimal damage to normal ones.
Key points
- •
The history of genetics and the Human Genome Project.
- •
Understanding genetics: from DNA to Protein.
- •
Identification of different molecular abnormalities.
- •
Most used technologies to identify genetic injuries.
- •
Genetics and epigenetics mechanisms identified in neurological disorders.
Introduction
Genome is the term used to describe the group of genes and regulatory sequences from an individual. Genes carry all the information that distinguishes one organism from others. Genes were discovered in 1865 by Gregor Mendel. His observations led to the creation of laws regarding the transmission of hereditary characteristics from generation to generation, which have constituted the basis of genetics until now. The nature of the genes was understood only in 1952, when the scientist Roselin Franklin showed a distinctive pattern that indicated the helical shape of DNA. One year later, James Watson and Francis Crick revealed the mechanisms of DNA structure: the double helix. In 1977, Frederick Sanger developed a rapid DNA sequencing technique.
In 1983, Kary Mullis improved the technique of PCR for amplifying DNA and the first genetic disease, Huntington disease, was mapped.
Since then, the development of techniques to analyze the genome has grown and different pathologies are associated with genetic abnormalities.
Recently, the DNA sequence of the entire human genome was sequenced by the international, collaborative research program called the Human Genome Project (HGP). The project was idealized in 1984 but launched in 1990. The full sequence of the human genome obtained by the HGP was completed in 2003 and provided the first complete view of human genetic code.
The complete human genome contains approximately 3 billion bases and approximately 20,500 protein-coding genes on 46 chromosomes (22 pairs of autosomal chromosomes and 2 sex chromosomes). The coding regions constitute less than 5% of the genome (the function of the remaining noncoding DNA is not yet well established) and some chromosomes have a higher density of genes than others. James Watson was one of the HGP heads and his own genome was sequenced and published on the Internet.
Scientists are studying how the DNA sequences of human genes can vary among individuals and populations and how genetic changes can generate diseases.
A genetic disease is any illness caused by an abnormality in an individual’s genome. The abnormality can range from a discrete mutation in 1 base in the DNA of a single gene to a chromosome aberration involving the gain or loss of the genes in an entire chromosome or set of chromosomes. Some genetic disorders are hereditary (inherited from the parents) whereas others are caused by acquired mutations in a somatic gene or group of genes. Mutations can happen either randomly or due to some environmental exposure.
Most genetic diseases are the direct result of mutations in 1 or many genes. One of the most difficult questions to be further elucidated, however, is how genes contribute to diseases that have a complex pattern of inheritance, such as diabetes, asthma, cancer, and mental illness.
In the nervous system, from depression to Alzheimer disease, familial genetic heritage has been observed. Other conditions, such as Parkinson disease and tumors in the central nervous system, have been associated with a variety of gene deregulations.
In these cases, more than 1 mutation is responsible for the disease arising, and several genes may contribute to a person’s susceptibility to a disease. Moreover, genes may affect how someone reacts to environmental factors. To understand how genetics are involved in the genesis of a disease, it is important to understand the mechanisms of gene expression, cell cycle, and proliferation/death control.
Introduction
Genome is the term used to describe the group of genes and regulatory sequences from an individual. Genes carry all the information that distinguishes one organism from others. Genes were discovered in 1865 by Gregor Mendel. His observations led to the creation of laws regarding the transmission of hereditary characteristics from generation to generation, which have constituted the basis of genetics until now. The nature of the genes was understood only in 1952, when the scientist Roselin Franklin showed a distinctive pattern that indicated the helical shape of DNA. One year later, James Watson and Francis Crick revealed the mechanisms of DNA structure: the double helix. In 1977, Frederick Sanger developed a rapid DNA sequencing technique.
In 1983, Kary Mullis improved the technique of PCR for amplifying DNA and the first genetic disease, Huntington disease, was mapped.
Since then, the development of techniques to analyze the genome has grown and different pathologies are associated with genetic abnormalities.
Recently, the DNA sequence of the entire human genome was sequenced by the international, collaborative research program called the Human Genome Project (HGP). The project was idealized in 1984 but launched in 1990. The full sequence of the human genome obtained by the HGP was completed in 2003 and provided the first complete view of human genetic code.
The complete human genome contains approximately 3 billion bases and approximately 20,500 protein-coding genes on 46 chromosomes (22 pairs of autosomal chromosomes and 2 sex chromosomes). The coding regions constitute less than 5% of the genome (the function of the remaining noncoding DNA is not yet well established) and some chromosomes have a higher density of genes than others. James Watson was one of the HGP heads and his own genome was sequenced and published on the Internet.
Scientists are studying how the DNA sequences of human genes can vary among individuals and populations and how genetic changes can generate diseases.
A genetic disease is any illness caused by an abnormality in an individual’s genome. The abnormality can range from a discrete mutation in 1 base in the DNA of a single gene to a chromosome aberration involving the gain or loss of the genes in an entire chromosome or set of chromosomes. Some genetic disorders are hereditary (inherited from the parents) whereas others are caused by acquired mutations in a somatic gene or group of genes. Mutations can happen either randomly or due to some environmental exposure.
Most genetic diseases are the direct result of mutations in 1 or many genes. One of the most difficult questions to be further elucidated, however, is how genes contribute to diseases that have a complex pattern of inheritance, such as diabetes, asthma, cancer, and mental illness.
In the nervous system, from depression to Alzheimer disease, familial genetic heritage has been observed. Other conditions, such as Parkinson disease and tumors in the central nervous system, have been associated with a variety of gene deregulations.
In these cases, more than 1 mutation is responsible for the disease arising, and several genes may contribute to a person’s susceptibility to a disease. Moreover, genes may affect how someone reacts to environmental factors. To understand how genetics are involved in the genesis of a disease, it is important to understand the mechanisms of gene expression, cell cycle, and proliferation/death control.
From DNA to protein
DNA is the molecule that carries hereditary information in almost all organisms. DNA consists of 2 polynucleotide strands. Each nucleotide comprises a sugar, a phosphate molecule, and a nitrogenous base (adenine, guanine, thymine, or cytosine). DNA is arranged in spiral forming a structure, called the double helix.
During DNA replication each strand acts as a template for the synthesis of its complementary strand. The disposition of nucleotides along the DNA strand constitutes the genetic code. Every three nucleotide sequence, called codon, encodes one specific amino acid. A gene is a sequence of nucleotides along the DNA strand that determines the sequence of amino acids in a protein.
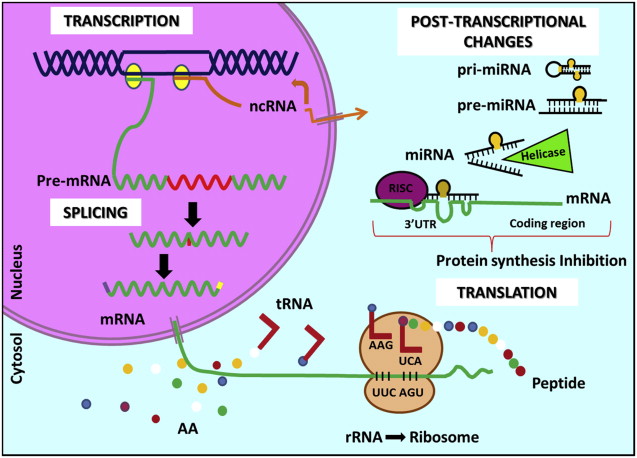
Gene expression is a process of DNA sequence reading into protein synthesis. This process has 2 major steps: transcription and translation. During transcription, the information is shifted from the DNA to a messenger RNA (mRNA). The DNA serves as template for complementary base pairing catalyzed by an enzyme called RNA polymerase, forming a precursor RNA molecule (pre-mRNA). Then the pre-mRNA is processed to form a mature mRNA. During this phase, called splicing, noncoding regions named introns are excluded from the molecule and only the coding region, the exons, remain in the mRNA structure. Alternative splicing occurs and different sequences are processed, giving rise to a huge variety of mature mRNAs. The mature mRNA receives some structural markers that signal for cytoplasmic exportation. In the cytoplasm, the step of protein synthesis called translation starts. The mature mRNA, a single-stranded copy of the gene, is then recruited by a protein complex in the ribosome and is translated into a protein molecule.
DNA Transcription—Control of Gene Expression
The process of transcription was first observed in 1970 by electron microscopy. During transcription, 1 DNA strand serves as template for RNA synthesis, whereas the other strand is considered noncoding.
The process of transcription starts when RNA polymerase attaches to the template DNA strand and begins to catalyze the production of RNA by matching complementary bases to the original DNA strand.
Transcription factors bind to specific DNA sequences called enhancer and promoter/silence sequences to recruit RNA polymerase to an appropriate transcription site and signal which part of the gene will be transcribed.
The promoters and enhancers or silencers are located within regulatory regions of the gene as well as within introns. Enhancer sequences regulate gene activation by binding proteins and changing the conformational structure of the DNA, helping to attract RNA polymerase. Because DNA is tightly packed into chromatin, transcription also requires several specialized proteins that facilitate the access to the coding strand.
The Role of Chromatin Structure
In human cells, DNA is packaged around histones, which are proteins in chromatin that also perform a function in gene regulation. In general, they control whether transcription factors may access the DNA.
For transcription to occur, the transcription area needs to be unrolled. This process requires the coordination of histone modifications, transcription factors binding, and other chromatin remodeling activities.
Modifications of histones can open up a gene, whereas DNA modifications can shut it down. In general, methylation of DNA makes chromatin more tightly closed and results in down-regulation or inhibition of gene transcription. On the other hand, acetylation of histones unbends bindings and helps transcription. Methylation of histones can increase or decrease their acetylation, regulating gene expression. The mechanism of regulation of DNA 3-D structure is called epigenetics.
DNA methylation is used in some genes to differentiate which copy is inherited from the father and which is inherited from the mother, in a process called imprinting. It can also help to explain why identical twins are not phenotypically identical. In addition, epigenetic control is responsible for X-chromosome inactivation in women that guarantees they have the same number of X-chromosome gene products as do men.
mRNA Processing (Splicing)
Splicing is the pre-mRNA processing mechanism by which introns are removed and exons are joined together to form the mature mRNA. Specific sequences in the pre-mRNA show where introns and exons are located. Splicing is usually constitutive, meaning that all exons are joined together in the same order they occur in the pre-mRNA. Alternative splicing is also observed, however, and the exons are combined in different ways. In alternative splicing, sequences may serve as exons and be included in the final mRNA, but under different conditions the same sequence can be treated as an intron and be removed from the mature mRNA.
Alternative splicing enables a single gene to generate more than 1 mRNA and contributes to the diversity of proteins in eukaryotes. Cells respond to environment modifications by changing gene expression and protein activity, which can also deregulate alternative splicing.
Scientists estimate that many human genetic diseases may involve splicing errors, making better understanding the splicing mechanisms an important area of research.
Other RNA Subtypes—The Noncoding RNA
Human DNA safely and stably stores genetic material in the nuclei of cells. In the meantime, mRNA carries the same information as DNA but is not used for long-term storage and can freely exit the nucleus. Although the mRNA sequence is complementary to the DNA template, it carries uracils instead of thymines and, after splicing, the molecule receives signals to cytoplasmic location.
Other types of RNAs are also transcribed, but they are differently processed and exported to the cytoplasm. Once they do not generate mRNA, they are referred to as bob-coding RNA, because they do not encode proteins. Two of them have been implicated to help mRNA translation:
- •
Transfer RNA (tRNA) carries the appropriate amino acids into the ribosome for inclusion in the new protein.
- •
Ribosomal RNA (rRNA) forms the ribosomes.
In addition to rRNA and tRNA, other noncoding RNAs exist in eukaryotic cells. These molecules participate in many essential functions. As a group, these RNAs are frequently referred to as small regulatory RNAs and are divided into different subgroups:
- •
Small nuclear RNAs (snRNAs) play a critical role in gene regulation by join in RNA splicing. snRNAs are found in the nucleus and are typically tightly bound to proteins in complexes called small nuclear ribonucleoproteins (snRNPs). The most abundant of these molecules are the U1, U2, U5, and U4/U6 particles, which are involved in splicing pre-mRNA to form mature mRNA.
- •
MicroRNAs (miRNAs) are small regulatory single-stranded RNAs that are approximately 22 to 26 nucleotides in length. Their functions in gene regulation were initially discovered in the nematode Caenorhabditis elegans. MiRNAs bind to the 3′ untranslated region of their target mRNAs through imperfect base pairing and inhibit translation. Therefore, miRNAs inhibit gene expression in a post-transcription phase. Additional studies indicate that miRNAs also play significant roles in cancer and other diseases.
- •
Small interfering RNAs (siRNAs) are another class of small RNAs that also inhibit gene expression. Specifically, 1 strand of a double-stranded siRNA molecule can be incorporated into a complex called RNA-induced silencing complex (RISC). This RNA-containing complex can then inhibit transcription of an mRNA that has a sequence complementary to its RNA component.
- •
Small nucleolar RNAs are abundant in nucleolar extracts. These molecules function to process rRNA molecules, often resulting in the methylation and pseudouridylation of specific nucleosides.
New forms of noncoding RNAs with novel functions continue to be discovered.
Protein Synthesis (Translation and Post-transcriptional Mechanisms of Control—Small Interfering RNA/MicroRNA)
The mature mRNA arrives at the cytoplasm and goes to the ribosome. There, the ncRNAs and a complex of enzymes are responsible for its translation into proteins. In the mRNA, each sequence of three letters (A, C, G, or U) is called a codon. Individual codons code for specific amino acids. After the machinery for translation is ready at the ribosome, tRNAs bring the amino acids and the peptide strands begins to be synthesized.
The presence of miRNAs and siRNAs can inhibit translation by two different mechanisms in association with a complex called RISC. The small interfering RNA (si-RNA) molecule is opened by the RNA-induced silencing complex (RISC) and remain bound to one single strand, which then binds to a sequence-specific region of the mRNA, signaling for another component of RISC, called Slicer, to cut the mRNA in the middle of the binding region. The cells do not recognize the cut mRNA and degrade it.
In the case of miRNA, an miRNA-induced silencing complex associates with the mature miRNA and binds to mRNA on the ribosome, blocking translation. MiRNAs can imperfectly bind complementary mRNA, as opposed to siRNAs, that require near-perfect binding.
One ncRNA has many different mRNA targets. These two mechanisms of post-transcriptional inhibition of gene expression by ncRNAs are being intensely studied and are associated with many different diseases. Scientists are trying to develop specific target drugs to deliver synthetic and antagonist ncRNAs to sick cells and combat specific pathologies ( Fig. 1 ).
Molecular abnormalities
In normal conditions during cell division, mistakes may happen in DNA replication. The cell has mechanisms to repair or eradicate the error (DNA repair enzymes). If the error is irrecoverable, the cell dies by apoptosis. In cases of problems in DNA repair/apoptosis mechanisms, cells accumulate genomic abnormalities and pass them through generations of cells. The accumulation of mutations causes diseases and even tumors, by changing transcription of genes or regulatory ncRNAs and by changing gene function.
In diseases like cancer, there are mutations in key genes that control normal cell processes, such as cell cycle, differentiation, and apoptosis. A person has two copies of each gene, one from the mother and one from the father. People with a hereditary predisposition to a disease inherit one copy of the damaged gene but the other copy is still normal. These people are considered heterozygous for the feature determined by that gene. Gene mutations that are inherited occur in the germline, meaning that all a person’s cells carry the mutation. During life, a mutation may occur in the other copy of the gene and the pearson starts to present a defective phenotype. Most mutations are not inherited, however; they are acquired by somatic cells during individuals’ lives (the mutations occur in one specific type of adult somatic cell, then not all cells carry the mutations; thus these cells cannot be transmitted to the descendants).
Examples of typical abnormalities include polymorphisms, chromosomes aberrations, point mutations that alter coding sequences, and changes on chromatin structure. These phenomena are explained as follows.
Single Nucleotide Polymorphisms
A single nucleotide polymorphism (SNP) is a DNA sequence variation occurring when a single nucleotide of a gene (A, T, C, or G) differs among individuals.
Within a population, SNPs can be referred as the allele of minor frequency, in other words, the less common variant carried by the part of population. SNPs are frequently expressed differently in human populations; therefore, an SNP allele that is common in one region or in one ethnic group may be rare in another.
SNPs may be found in coding and noncoding regions of gene or in the intergenic regions between genes. SNPs within a coding sequence may not change the amino acid sequence due to degeneracy of the genetic code (both sequences led to the same polypeptide). SNPs that change the amino acid in general lead to different phenotypes but do not cause problems to the cell. SNPs that are not in protein coding regions may still have consequences for gene splicing, transcription factor binding, or the sequence of ncRNAs, also implying in change of protein synthesis.
SNPs occur in more than 1% of the population. They are common enough to be considered a normal genetic variation and are responsible for many of the normal differences between people, such as eye color, hair color, and blood type. Although most SNPs do not have negative effects on a person’s health, some of them may influence the risk of developing diseases.
Chromosomal Aberrations
There are 2 main types of chromosomal abnormalities: numeric and structural aberrations. Any increases or decreases in chromosomal material interfere with normal development and function.
- •
Structural aberrations
Structural aberrations can be intra- or interchromosomal. Intrachromosomal aberrations include deletion, duplication, and inversion. Interchromosomal include translocations. Deletions can happen naturally or be caused by chemical mutagens and radiation. When part of a dominant allele is deleted, it may cause the expression of a recessive character, configuring pseudodominance.
In turn, duplications occur in a lower frequency than deletions. In inversion, a segment of chromosomes is inverted on reversed by 180°. Translocations involve two nonhomologous chromosomes and position of part of the chromosome is changed, leading to change in arrangement of chromosomes.
Alteration in chromosome structure has been associated with prognosis and therapy response of many diseases, for example, translocations found in leukemias and lymphomas. Almost all patients with chronic myeloid leukemia carry the fusion gene BCR-ABL , which results from the translocation of chromosome 9 and 22. In solid tumors, individual structural aberrations have not been shown sufficiently prevalent, except for prostate cancer, in which 70% of the patients carry the fusion gene TMPRSS2 – ETS .
- •
Copy number aberrations
Numeric aberrations are caused by a defect in chromosome division, resulting in cells with extra chromosomes or a deficiency in chromosomes.
Gametes with these anomalies can result in conditions, such as Down syndrome (47 chromosomes instead of 46) or Turner syndrome (45 chromosomes).
Many regions of amplification and lower-level copy number abnormalities have been identified by comparative genomic hybridization (CGH) in different cancers, for example, NMYC in euroblastoma, EGFR in glioblastoma, and RB1 in retinoblastoma.
Point Mutations, Insertions, Deletions, and Duplications
The DNA sequence of a gene can be altered in several ways. Gene mutations have varying effects on health, depending on where they occur and whether they alter the function of resulting proteins. The types of mutations include
- •
Missense mutation—changes 1 DNA base pair and results in protein changes
- •
Nonsense mutation—also changes 1 DNA base pair but instead of changing 1 codon for another, the altered sequence signals to stop transcription, resulting in a shortened protein
- •
Insertion—addiction of bases in the DNA sequence, resulting in a protein that may not work properly
- •
Deletion—loss of a piece of the DNA sequence; small deletions remove 1 or a few base pairs from a gene, and larger deletions remove an entire gene or nearby genes
- •
Duplication—part of the DNA sequence is abnormally copied 1 or more times
- •
Frameshift mutation—when the addition or loss of DNA bases changes a gene’s reading frame. A reading frame consists of groups of 3 bases that each code for 1 amino acid. A frameshift mutation shifts the grouping of these bases and changes the code for amino acids. The protein resultant is often nonfunctional. Insertions, deletions, and duplications can all be frameshift mutations.
Epigenetics
Epigenetic silencing is 1 way to turn genes off, and it can contribute to differential protein expression among cells from different tissues. Epigenetics is involved in many normal cellular processes but it has been associated with many diseases. There are 3 epigenetic systems that can interact with each other to silence genes: DNA methylation, histone modifications, and RNA-associated silencing.
In DNA methylation, a methyl group is added to DNA. It is highly specific and always happens in a region in which a cytosine is located next to a guanine linked by a phosphate (called a CpG site). CpG sites are methylated by 1 of 3 enzymes called DNA methyltransferases. Inserting methyl groups changes the appearance and structure of DNA, modifying a gene’s interactions with the machinery within a cell’s nucleus that is needed for transcription.
When histones are modified after they are translated into protein, they settle chromatin arrangement, which can determine whether the associated DNA is transcribed. If chromatin is opened, DNA can be transcribed. But if chromatin is condensed (creating a complex called heterochromatin), DNA is not transcribed.
There are 2 ways histones can be modified: by acetylation and by methylation. Acetylation is usually associated with active chromatin, whereas deacetylation is associated with heterochromatin. On the other hand, histone methylation can be a marker for both active and inactive regions of chromatin.
Genes can also be turned off by RNA-associated silencing when it is in the form of antisense transcripts, ncRNAs. RNA might affect gene expression by causing heterochromatin to form or by initiating histone modifications and DNA methylation.
Epigenetic changes are required for normal processes but they can also be involved in some mechanisms of pathogenesis. Disrupting any system that contributes to epigenetic alterations can cause abnormal activation or silencing of genes. Such disruptions have already been associated with cancer, syndromes involving chromosomal instabilities, and mental retardation.
Genome Instability and Loss of Heterozygosity
When the normal mechanisms of epigenetic control are altered, genome instability is lodged in the cell. Genome instability causes nondisjunction during mitosis, segregation during recombination, or deletion of a chromosome segment. This phenomenon provides conditions for the loss of 1 copy of a gene allele (or loss of heterozygosity [LOH]) due to the accumulation of mutations. LOH becomes critical when the remaining allele contains a point mutation that changes protein expression. Then, people who already carry heterozygous mutations in 1 allele of an important gene may have the other allele lost or inactivated. This is a common occurrence in cancers where a tumor suppressor gene is affected.
Telomere Maintenance Mechanism
Telomerase is a ribonucleoprotein that elongates telomeric DNA by adding hexanucleotide repeats to compensate for progressive DNA loss that happens during each cell division. Telomerase consists in a catalytic protein subunit with reverse transcriptase activity (h-TERT), encoded by the hTERT gene and an RNA component (TERC), encoded by TERC gene, that serves as a template for the telomere repeat. Recently, high-expression levels of TERC and hTERT were associated with worse survival of children with non-brainstem high-grade gliomas, making telomerase a promising potential therapeutic target in this disease ( Fig. 2 ).