Glioblastoma (GBM) is the most common and most aggressive primary malignant tumor of the central nervous system. Recently, researchers concluded that the “one-size-fits-all” approach for treatment of GBM is no longer valid and research should be directed toward more personalized and patient-tailored treatment protocols. Identification of the molecular and genomic pathways underlying GBM is essential for achieving this personalized and targeted therapeutic approach. Imaging genomics represents a new era as a noninvasive surrogate for genomic and molecular profile identification. This article discusses the basics of imaging genomics of GBM, its role in treatment decision-making, and its future potential in noninvasive genomic identification.
Key points
- •
Glioblastoma constitutes a major challenge being the most aggressive and therapy-resistant malignant brain tumor.
- •
Personalized targeted treatment protocols are the focus of research as underlying genomic and molecular pathways are continuously being identified.
- •
Imaging genomics represents a new branch in science that links currently used imaging modalities to predict and correlate genomic profiles in glioblastoma tumors noninvasively.
Glioblastoma, overcoming the challenge
Glioblastoma (GBM; World Health Organization [WHO] grade IV astrocytoma) is the most common malignant primary brain tumor accounting for 45.2% of malignant primary brain tumors and for about 15.6% of all primary brain tumors. The best current standard of care for GBM includes surgical resection followed by adjuvant local radiotherapy and systemic chemotherapy with temozolomide (TMZ). Despite this aggressive multimodal treatment, the relative survival estimates for GBM are low; less than 5% of patients survived 5 years postdiagnosis. The median overall survival duration for patients ranges from 12.2 to 15.9 months. This is thought to be attributed to the highly infiltrative nature and the heterogeneity that GBM exhibits on molecular and genomic levels, which lead to differences in individual treatment response and prognosis. Approximately 90% of GBM tumors recur at the site of surgical resection because of invasive tumor cells that were left behind after resection and managed to avert the radiation therapy.
Although considered one histologic group, high-grade gliomas exhibit genetic and molecular heterogeneity to the extent that the “one-size-fits all” model for treatment of GBM is no longer valid; thus, research is now being directed toward a more personalized approach in treatment of patients with GBM. Establishment of this personalized treatment approach requires thorough understanding of the disease processes and underlying genomic and molecular pathways in the development of GBM along with identification of molecular features of the tumor that can predict response so that selection of patients who are most likely to benefit from a particular treatment can be done.
Glioblastoma, overcoming the challenge
Glioblastoma (GBM; World Health Organization [WHO] grade IV astrocytoma) is the most common malignant primary brain tumor accounting for 45.2% of malignant primary brain tumors and for about 15.6% of all primary brain tumors. The best current standard of care for GBM includes surgical resection followed by adjuvant local radiotherapy and systemic chemotherapy with temozolomide (TMZ). Despite this aggressive multimodal treatment, the relative survival estimates for GBM are low; less than 5% of patients survived 5 years postdiagnosis. The median overall survival duration for patients ranges from 12.2 to 15.9 months. This is thought to be attributed to the highly infiltrative nature and the heterogeneity that GBM exhibits on molecular and genomic levels, which lead to differences in individual treatment response and prognosis. Approximately 90% of GBM tumors recur at the site of surgical resection because of invasive tumor cells that were left behind after resection and managed to avert the radiation therapy.
Although considered one histologic group, high-grade gliomas exhibit genetic and molecular heterogeneity to the extent that the “one-size-fits all” model for treatment of GBM is no longer valid; thus, research is now being directed toward a more personalized approach in treatment of patients with GBM. Establishment of this personalized treatment approach requires thorough understanding of the disease processes and underlying genomic and molecular pathways in the development of GBM along with identification of molecular features of the tumor that can predict response so that selection of patients who are most likely to benefit from a particular treatment can be done.
The Cancer Genome Atlas and genetic biomarker identification
Atkinson and colleagues defined a biomarker as “a characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathogenic processes, or pharmacologic responses to a therapeutic intervention.” Predictive biomarker compares the effect of a certain intervention for marker-positive versus marker-negative patients and predicts the differential effect of this intervention on the outcome (ie, indicate whether the patient is likely or unlikely to benefit from a specific drug or regimen). Prognostic biomarker provides information about the patient’s probable long-term outcome (overall survival), either untreated or with a standard treatment.
With the change in perception of cancer as a disease of genetic alterations, it has been concluded that the time interval between initial development of malignancy and the initial diagnosis and intervention allows multiple genetic alterations to accumulate. Accordingly, most cancers now are associated with change in the expression of multiple genes and not just a single gene. This diversity in genetic alterations renders identification of certain predictive and prognostic genetic biomarkers for GBM a great challenge. During the last decade, the use of microarray-based high-output sequencing methods that allowed simultaneous measurement of many genes and their products, and further quantitative analysis of the entire gene networks has deepened the insight of the processes underlying the development of GBM.
Thorough analysis of the Cancer Genome Atlas (TCGA) data concluded that GBM harbors more than 60 genetic alterations including genetic mutations and chromosomal aberrations. TCGA, which began in 2006, is a publicly available large-scale multi-institutional collaborative effort funded by the National Cancer Institute and the National Human Genome Research Institute to establish a better understanding of the molecular alterations of cancer associated with pathologic and radiologic features and response to therapy. GBM was the first cancer examined by TCGA. By identification of specific genetic mutations and microRNA expressions in nearly 500 tumor samples, TCGA-driven studies increased awareness of the significance of the genetic and molecular pathways in the pathogenesis of GBM and the identification of potential novel molecular therapeutic targets. Typical identified alterations included mutations affecting epidermal growth factor receptor ( EGFR ), tumor protein 53 ( TP53 ), cyclin-dependent kinase inhibitor 2a, cyclin-dependent kinase 4, retinoblastoma 1, and phosphatase and tensin homolog ( PTEN ) genes and deletions on several chromosomal arms including 1p, 9p, 10p, 10q, 13q, 17p, 19q, and 22. Some of the most significant GBM genetic biomarkers that have been identified include EGFR , O 6 -methylguanine–DNA methyltransferase ( MGMT )-promoter methylation, isocitrate dehydrogenase-1 ( IDH1 ) mutation, 1p/19q co-deletion, and TP53 mutation.
Molecular classification of glioblastoma
This breakthrough in genetic identification paved the path for explaining the difference in survival and response to treatment among patients of GBM through replacing the classical classification of GBM with another molecularly based classification. Classically, GBM was classified into primary GBM (which presents at the time of diagnosis as advanced cancers with no previous evidence of lower-grade lesion) and secondary GBM (those who have clinical, radiologic, or histopathologic evidence of malignant progression from a pre-existing lower-grade tumor). Based on the differences in genetic aberrations and gene-expression, further classification of GBM identified four molecular subtypes: classical, mesenchymal, proneural, and neural, which are different in gene expression of EGFR , neurofibromin 1 ( NF1 ), alpha-type platelet-derived growth factor receptor ( PDGFRA/IDH1 ), and ERBB2 , respectively. Patients of the classical and mesenchymal subtypes of GBM showed significantly reduced mortality and improved survival with the use of aggressive concurrent chemoradiotherapy. However, survival was not altered in the proneural subtype. Molecular analysis of low-grade gliomas is currently underway, and it is anticipated that the data will provide similar molecular subclassification of low-grade gliomas.
Principal genetic biomarkers in glioblastoma
It is important that radiologists understand common genetic alterations. Certain imaging features of GBMs may correlate with certain gene expressions. Imaging can provide a noninvasive technique to assess spatial and temporal changes in gene expression. Understanding the molecular changes that occur in GBMs is helpful in explaining imaging findings in patients undergoing molecular targeted therapies.
TMZ, an alkylating therapeutic agent used in treatment of GBM, acts through methylation of the O 6 position of guanine, thus triggering cytotoxicity and apoptosis of cancer cells. Although O 6 -alkylguanine is not the main lesion induced by alkylating agents, it seems to be the most cytotoxic one. Cellular protection against the effect of such alkylating agents is mediated through the MGMT gene, located on chromosome 10q26, which encodes for a DNA repair protein (MGMT) that is responsible for removing the alkyl groups from the O 6 position of guanine. Epigenetic slicing of MGMT gene by promoter methylation suppresses the gene expression, leading to reduction of the amount of MGMT protein and subsequently reduces the DNA repair capacity of the cell. This renders the cell more prone to the action of alkylating therapeutic agents because of the accumulation of O 6 -alkylguanine, which subsequent to incorrect pairing with thymidine triggers mismatch repair, thereby inducing DNA damage and, eventually, cell death. Cancer cells harboring higher levels of MGMT gene expression, which blunts the effect of alkylating therapeutic agents, are considered to be a treatment-resistant group and patients belonging to this group are less likely to benefit from treatment with alkylating agents. Accordingly, MGMT -promoter methylation status is considered to be the strongest prognostic factor for outcome in patients with newly diagnosed GBM, and is a powerful predictor of response to alkylating chemotherapy.
IDH enzyme plays an important role in the cellular protection against oxidative damage. IDH exists in three isoforms: IDH1, which is strictly cytosolic, and IDH2 and IDH3, which are located inside the mitochondria. These enzymes are responsible for conversion of isocitrate to α-ketoglutarate, a process that is accompanied by reduction of NADP + into NADPH (NAD + into NAD in case of IDH3). Only mutations of IDH1 and IDH2 were found in high proportions in gliomas, with IDH1 mutations being much more common than IDH2 . In fact, gliomas are considered to be the most likely cancer type to show IDH gene mutation. Altered IDH1 enzyme found in tumor cells consists of dimer between the wild and the mutated enzymes. Mutated IDH1 enzyme contributes to the pathogenesis of gliomas through different mechanisms, most prominent of which is reduction of α-ketoglutarate into 2-hydroxyglutarate resulting in inhibition of the activity of a variety of dioxygenases (eg, prolyl hydroxylase), with subsequent activation of hypoxia-inducible factor (HIF)-1α transcription factor and promotion of vascular endothelial growth factor (VEGF)–mediated angiogenesis, eventually leading to greater tumor growth. IDH1 mutations have shown to be an independent prognostic biomarker in patients with glioma. Patients with mutated IDH1 have shown a better prognosis (5-year survival rate, 93%) than those without the mutation (5-year survival rate, 51%) in terms of overall and progression-free survival. Recent studies have showed that IDH1 mutation also can have a potential predictive role. In secondary patients with GBM, IDH1 mutations and MGMT -promoter methylation status were strongly correlated with increased overall survival. Patients with both IDH1 mutation and MGMT -promoter methylation had the best response rate to TMZ (median progression-free survival, 13.4 months) followed by patients with IDH1 mutation alone (10.2 months), and finally patients with none of these genetic alterations had the worst response rate to TMZ (6.1 months). Because of its value as a biomarker, IDH1 mutation status is likely to result in revisions to the current WHO classification scheme for GBM.
VEGF is one of the most critical proangiogenic factors that play an integral role in brain angiogenesis. The VEGF gene contains a hypoxia-responsive element within its promoter that binds HIF-1α, thereby activating transcription. VEGF is responsible for directing nearby angiogenesis. It was proved that inhibition of this HIF/VEGF pathway suppresses tumor growth experimentally. Once expressed and secreted, extracellular VEGF binds to its high-affinity receptors, VEGFR-1 and VEGFR-2, which are upregulated on endothelial cells of high-grade gliomas, but not present in normal brain. Receptor activation then leads to angiogenesis in regions adjacent to tumor cells leading to the high vascularity noted in GBM tumors.
EGFR is a transmembrane tyrosine kinase receptor. EGFR gene overexpression is found in more than 50% of patients with GBM, and EGFR amplification and mutation are strongly associated with GBM transformation. EGFR normally exists as a monomer that transforms into homodimers on binding to EGF with consequent kinase activation and autophosphorylation. This leads to activation and phosphorylation of EGFR variant III (EGFRvIII). EGFRvIII is found in a subgroup of patients (20%–40%) who undergo EGFR gene rearrangements and it lacks the extracellular domain. Activated EGFR stimulates EGFRvIII resulting in activation of downstream signaling pathways, such as receptor tyrosine kinase, phosphoinositide 3-kinase, and PTEN eventually leading to augmentation of tumor angiogenesis, cell proliferation, and cell survival. Heimberger and colleagues found that EGFRvIII has a negative prognostic value in patients surviving more than 1 year. Therapies have been developed that are directed at the overexpressed EGFR in GBM. Multiple studies demonstrate how EGFR status affects GBM response to treatment. Mellinghoff and colleagues demonstrated that GBMs characterized by coexpression of EGFRvIII and PTEN yield the best response to therapy using EGFR inhibitors. Accordingly, it might become important to determine the EGFR status of patients with GBM before therapy to stratify patients into clinical trials and also to predict which group of patients would benefit most of the treatment.
1p/19q loss of heterozygosity (LOH) is the genetic hallmark of oligodendrogliomas and anaplastic oligodendrogliomas (WHO grades II and III, respectively). It is found in 40% to 90% of the patients with oligodendrogliomas and it is linked to longer survival. It also predicts a favorable response to chemotherapy and radiotherapy. However, in GBM, the prognostic value of these alterations is still debatable. In GBM, 1p/19q codeletion is primarily associated with GBM with oligodendroglial component (GB-O). The percentage of GB-O cases in GBM population ranges from 4.2% to 27.2%. The presence of 1p/19q codeletion was observed in 22% to 28% of GB-O patients, whereas LOH 1p was observed in 24% of patients with GBM and LOH 19q was detected in 5% to 33% of patients with GBM. However, the impact of LOH 1p on survival is unclear and the results of previous studies are inconsistent. Homma and colleagues studied 209 patients with GBM. A total of 21% of these patients were LOH 1p and/or 19q positive. LOH 1p patients showed longer survival than patients without LOH 1p (13.2 ± 10.8 vs 9.6 ± 7.4 months; P = .0536). After age and gender adjustment, multivariate analysis showed longer survival in patients with LOH 1p (hazard ratio, 0.7; 95% confidence interval, 0.5–1.0).
Basics of imaging of glioblastoma
Histopathologic Imaging
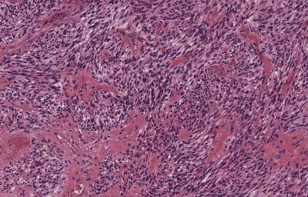
Despite the diversity of appearances, the single most important pathologic characteristic of GBM that discriminates it from lower-grade astrocytomas is the presence of central necrosis ( Fig. 1 ). GBM is also characterized by the presence of significant necrosis, microvascular proliferation, and invasion in addition to other feature of anaplastic astrocytomas including cellularity, nuclear atypia, and mitotic activity. GBM is also characterized by “pseudopalisading,” in which areas of viable neoplastic cells form an irregular border surrounding areas of necrotic debris. This characteristic pathology reflects the basis of the imaging findings of GBM. Out of all available imaging modalities, MR imaging stands as the imaging modality of choice for diagnosis, pretreatment surgical planning, and posttreatment monitoring of GBM.
MR Imaging
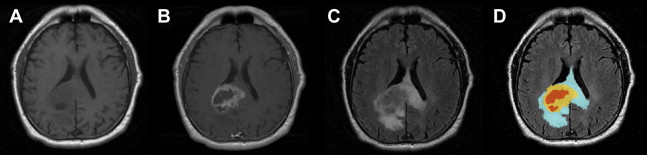
MR imaging techniques of GBM are shown in Table 1 . Classically, GBM is described on MR imaging as a thick irregular ring of heterogeneous enhancement surrounding a central nonenhancing core of necrosis and surrounded by an area of peritumoral hyperintensity, the latter more evident on FLAIR and T2-weighted sequences, representing the region of tumor infiltration and vasogenic edema ( Fig. 2 ). In fact, neoplastic cells extend beyond the detected imaging changes and in areas of normal-appearing white matter.
| Conventional Techniques | Advanced Techniques |
|---|---|
| T1-weighted imaging | MR perfusion imaging |
| T2-weighted imaging | MR spectroscopy imaging |
| FLAIR imaging sequence | MR diffusion-weighted imaging |
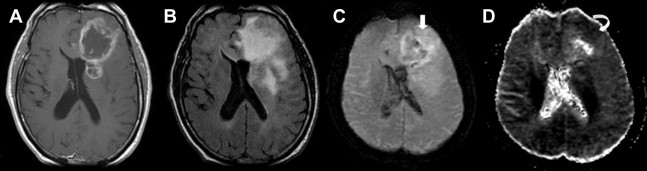
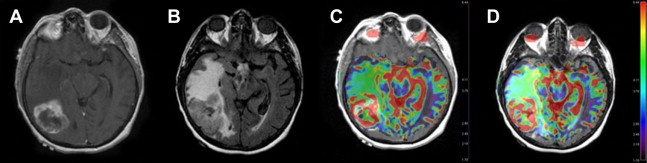
Advanced imaging techniques including magnetic resonance (MR) diffusion, perfusion, and spectroscopy enhance the anatomic data supplied by conventional MR imaging and provide physiologic information, such as cellularity, microperfusion, and metabolism, respectively. Diffusion-weighted imaging measures the diffusion of water molecules in a voxel. In GBM, diffusion is restricted in areas with a high density of cell packing/tumor cells and high nuclear-to-cytoplasm ratios ( Fig. 3 ). Findings on diffusion-weighted echoplanar MR images can be used to distinguish nonenhancing tumor from peritumoral edema when these abnormalities are located in white matter and to differentiate various components of tumor (enhancing, nonenhancing, cystic, or necrotic). MR perfusion imaging assesses angiogenesis and blood-brain barrier permeability, which can be predictive of the tumor grade and higher malignant histology ( Fig. 4 ). Moreover, as novel therapeutic agents targeting angiogenesis in brain tumors are being developed, the role of perfusion imaging as an important tool for diagnosis and follow-up becomes more evident. Lastly, MR spectroscopy measures the changes in metabolites in the region of interest based on chemical information obtained from the MR signal.
These characteristic imaging findings correspond to the unique pathologic features that GBM exhibits. Interestingly, regional genetic and cellular expression patterns of GBM were found to influence the anatomic and physiologic MR imaging. This significant relationship plays an important role in the potential value of the use of MR imaging as a noninvasive surrogate for identification of these genetic alterations.
Imaging genomics: a new dimension for diagnostic imaging
Genetic profiling of GBM as the basis of the personalized treatment approach necessitates obtaining tissue specimens from the tumor with subsequent histologic and immunohistochemical analysis. Accordingly, obtained results depend on the surgical procurement of the specimen, the part of the tumor from which this specimen biopsy was obtained, and the analysis methods used for genetic identification. Because of the heterogeneity of genetic expression that GBM exhibits in between the different tumor components, the results obtained from the tumor sample only reflect the genetic alterations in this part of the tumor and not the tumor as a whole, leading to inaccurate results. Moreover, this technique, being invasive, carries a lot of risks and drawbacks. It involves the risk of pain, infection, hemorrhage, and the complications associated with any surgical procedure. Also it is not always possible to obtain a sample of the tumor especially if it is located in a risky or inaccessible location of the brain tissue. It became clear that this technique for genetic profiling is unsuitable for routine use for every patient with GBM. Consequently, the need for a safer, noninvasive, accurate, and comprehensive alternate for genetic identification has drawn attention to other techniques that can be routinely used with minimal risks and complications. MR imaging of the brain has been demonstrated to have a strong potential as a surrogate for GBM genetic profiling. This interconnection between radiologic imaging and genomics is described by the term “imaging genomics.”
Imaging genomics is a new field in science that bidirectionally links specific imaging traits, better called radiophenotypes, to genomic profiles thus providing a novel bridge between two entities, namely imaging and genomics, that until recently had not been well-elucidated. In imaging genomics imaging traits obtained from routine imaging studies are being associated with certain gene expression patterns so these traits can be used as a noninvasive surrogate for providing molecular information about the tumor’s genetic profiles. This may further strengthen the integration of multiple data ( Fig. 5 ).