9
MOLECULAR TECHNIQUES, ASSAY SYSTEMS, AND SIGNALING IN RADIOBIOLOGY
KAILIN YANG AND JENNIFER S. YU
Question 1
What is cytogenetics and how is a cytogenetic study performed?
Question 1 What is cytogenetics and how is a cytogenetic study performed?
Answer 1
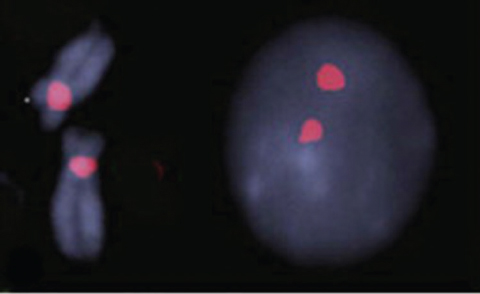
Cytogenetics is the study of chromosome number and structure. The first step of a cytogenetic study is the preparation of chromosome spreads by mechanically rupturing metaphase cells that are hypotonically swollen. The chromosomes are spread across a small region on a microscope slide and stained. These stained chromosomes are individually recognized and classified. Traditional Q-banding used alkylating fluorochromes to stain chromosomes. Modern banding techniques can generate 300 to 850 bands across all chromosomes of the human genome. Traditional cytogenetics can be complemented with fluorescence in situ hybridization (FISH) and genomic microarray analysis (Figure 9.1).
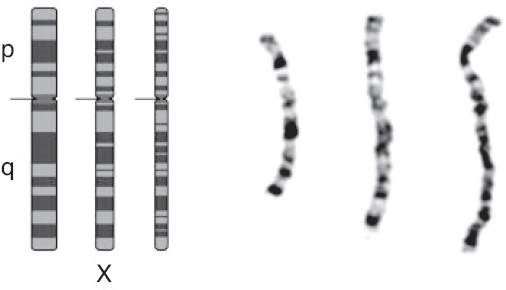
Figure 9.1 Ideograms and photomicrographs of the X chromosome (from left to right: metaphase, prometaphase, and prophase).
Source: Figure 9.1 from Nussbaum RL, McInnes RR, Willard HF. Thompson & Thompson Genetics in Medicine. 8th ed. Philadelphia; PA: Elsevier, 2016:57–74.
Spellman PT, Costello JF, Gray JW. Cancer genomics. In: Mendelsohn J, Howley PM, Israle MA, Gray JW, Thompson CB, eds. The Molecular Basis of Cancer. 3rd ed. Philadelphia, PA: Saunders/Elsevier; 2008:267–282.
Question 3
What is the basic mechanism of Sanger sequencing?
Question 3 What is the basic mechanism of Sanger sequencing?
Answer 3
Sanger sequencing is the mainstay of traditional sequencing methodology used to detect mutations in a specific gene. The DNA template to be sequenced is subjected to a modified version of the polymerase chain reaction (PCR): standard deoxynucleotides (dNTPs: dATP, dTTP, dCTP, dGTP), DNA primer, and DNA polymerase are added to the DNA template, and the mixture is divided into four separate PCRs. To each reaction, a different fluorescently labeled dideoxynucleotide (ddATP, ddTTP, ddCTP, ddGTP) is added at a low concentration to terminate the DNA elongation. The DNA population of each modified PCR reaction is a mixture of DNA fragments of different lengths labeled with a fluorescent dideoxynucleotide at one end. Electrophoresis is then performed for each of the four DNA populations, to generate a chromatogram to decode a full sequence of the original DNA template (Figure 9.3).
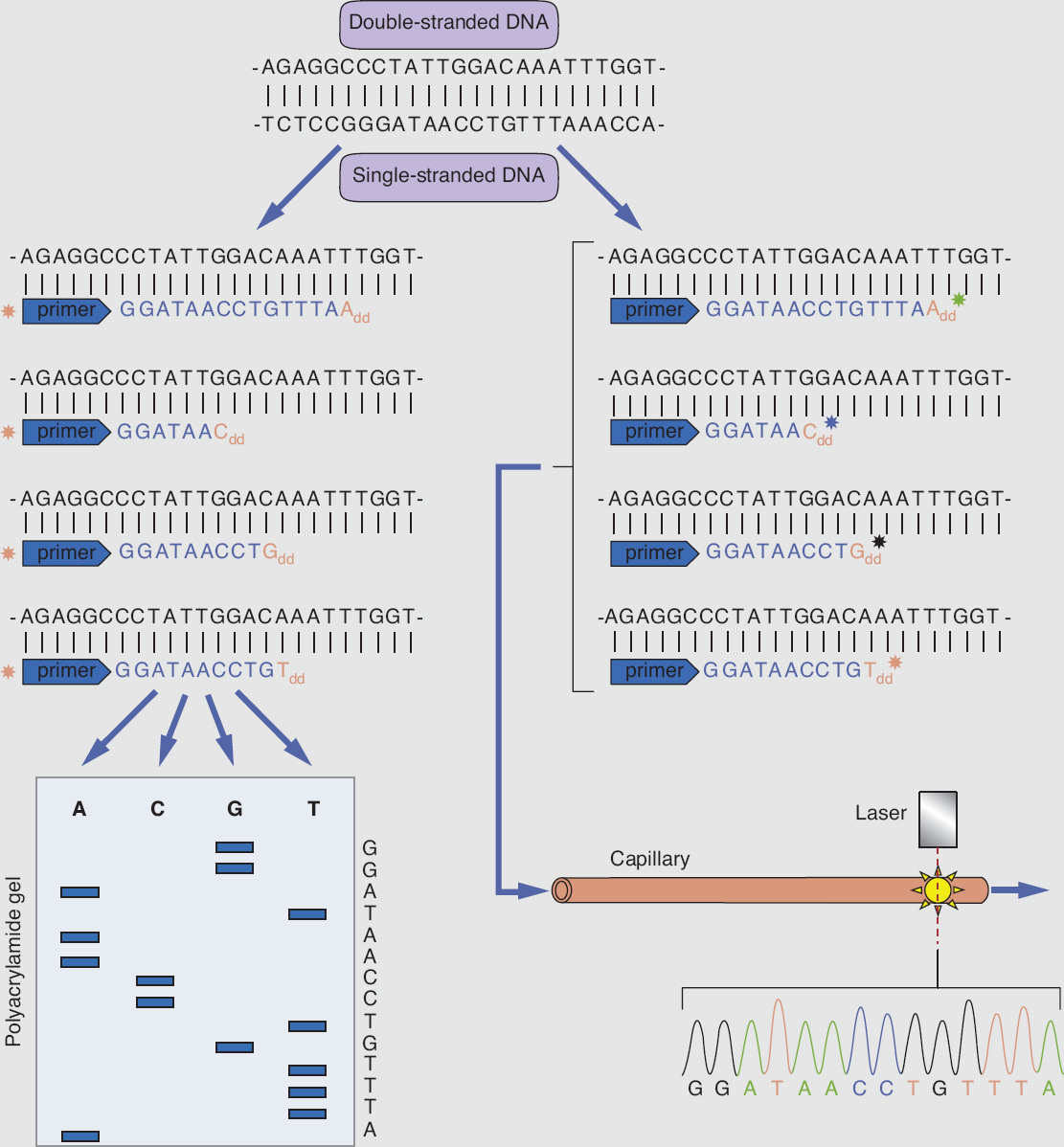
Figure 9.3 Schematic presentation of Sanger DNA sequencing, using chain termination method.
Source: Figure 9.3 from Pitt AR, Kolch W. Medical Biochemistry. 4th ed. Philadelphia, PA: Saunders/Elsevier; 2014:466–485.
Spellman PT, Costello JF, Gray JW. Cancer genomics. In: Mendelsohn J, Howley PM, Israle MA, Gray JW, Thompson CB, eds. The Molecular Basis of Cancer. 3rd ed. Philadelphia, PA: Saunders/Elsevier; 2008:267–282.
Question 4
What is DNA methylation? What methods have been developed to study DNA methylation status in cancer cells?
Question 6
What is the phenotypical consequence of congenital mutations impairing the function of a DNA damage response gene?
Question 4 What is DNA methylation? What methods have been developed to study DNA methylation status in cancer cells?
Answer 4
DNA methylation is defined as the addition of methyl group onto DNA itself, specifically to the cytosine base to form 5-methylcytosine. Such cytosine is usually followed with guanine at its 3′ end, forming a CpG island for methylation. This reaction is catalyzed by DNA methyltransferase. Cells use DNA methylation as a form of epigenetic regulation on gene expression. In cancers, an aberrant pattern of DNA methylation modulates gene expression to promote tumorigenesis and resistance to treatment. Several methodologies have been developed to detect DNA methylation, including methylation-specific PCR at a single gene, restriction-landmark genomic scanning, microarray-based DNA methylation analysis, and reduced-representation bisulfite sequencing. The method of reduced-representation bisulfite sequencing uses a combination of restriction enzymes and bisulfite sequencing to detect DNA methylation on the genome. Bisulfite treatment of DNA converts unmethylated cytosine nucleotide into uracil, while 5-methylcytosine is not affected. Sophisticated bioinformatics analysis is then performed to interpret the methylation status based on the sequencing results from bisulfite-treated DNA samples.
Spellman PT, Costello JF, Gray JW. Cancer genomics. In: Mendelsohn J, Howley PM, Israle MA, Gray JW, Thompson CB, eds. The Molecular Basis of Cancer. 3rd ed. Philadelphia, PA: Saunders/Elsevier; 2008:267–282.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree