Fig. 1
Sequence logos for classical and selective AREs. Classical AREs resemble inverted repeats of the 5′-AGAACA-3′-like motif spaced by three nucleotides, while selective AREs resemble direct repeats of the same motif. Logos were created on the weblogo.berkeley.edu website, based on a list of all published AREs for which selectivity was checked in electrophorectic mobility shift assays and in functional analysis
2.2 Selective AREs
The DBD of AR, GR, PR, and MR are very similar, with identity of the residues involved in contacting the DNA and high similarity of the dimerization interface. However, since each corresponding hormone has its specific target genes, even in cells where the receptors are co-expressed, efforts have been made to find DNA sequences that are selective for any of the four receptors. In vitro DNA motif selections, based on PCR amplifications of DBD-bound oligonucleotides, did not reveal selective elements (Roche et al. 1992) . It was only through the analysis of a series of AREs isolated from androgen target genes that it became apparent that several of these AREs were not recognized by the GR-DBD. These so-called selective AREs consist of a 5′-AGAACA-3′-like hexamer, flanked at three nucleotides downstream by a second hexamer. The similarity of this downstream hexamer to the 5′-AGAACA-3′ consensus is lower compared to that in the classical AREs. In fact, initial mutation analyses indicated that selective AREs resemble a direct, rather than a palindromic, repeat of the 5′-AGAACA-3′ consensus (Claessens et al. 1996; Verrijdt et al. 2000) (Fig. 1) .
For the DBD of the ER, residues in the first zinc finger module dictate higher affinity for 5′-AGGTCA-3′, while in GR, AR, PR and MR, alternative residues at the same positions dictate high affinity for 5′-AGAACA-3′ (Umesono et al. 1989) . Since the two hexamers that constitute all AREs , both classical and selective, resemble the same consensus, it is not surprising that the binding of the AR to selective AREs and the non-binding of the GR to these elements is not determined by differences in the first zinc finger. However, a chimeric AR in which the second zinc finger of the DBD is replaced by that of the GR possesses significantly attenuated affinity for selective AREs in transient transfections experiments and in vitro binding assays (Schoenmakers et al. 1999) . This finding indicates that the second zinc finger of the AR allows dimerization on selective elements, while the second zinc finger of the GR does not .
2.2.1 In vivo Role
Based on the in vitro data on the role of the second zinc finger in selective ARE binding, and the fact that this receptor fragment is encoded by a separate exon in the AR as well as in the GR gene, a transgenic model was developed in which this exon in the AR gene was swapped by that of the GR gene. The resulting model, called SPARKI for ‘SPecificity affecting AR Knock In’ expresses an AR that still binds classical AREs with high affinity but has lost high affinity for selective AREs (Schauwaers et al. 2007) . In effect, this model can be considered a knockout of selective AREs . These mice have smaller reproductive organs and reduced fertility, while no differences are observed in other androgen target tissues like bone, muscle, kidney or lacrimal glands. Therefore, it seems that selective AREs are not involved in the anabolic effects of androgens, but have a specific role in reproduction.
The reduced fertility observed in SPARKI is mainly explained at two sites: in the testis, the number of Sertoli cells is reduced and the spermatogenic process seems to be affected at the second meiotic division; in the epididymis, the sperm maturation is impaired and this correlates with the reduced expression of a subset of the androgen-regulated genes in this tissue. Several of these genes have a known role in sperm maturation, and selective AREs were identified in two of them which is proof for the fact that the second zinc finger of the AR has a crucial role in the recognition of selective AREs (Kerkhofs et al. 2012) .
Several of the AREs described in AR-binding segments found in human prostate cancer cell lines are selective AREs (Denayer et al. 2010; Wang et al. 2007) . The fact that the prostate of SPARKI mice is smaller (Schauwaers et al. 2007) indicates that selective AREs have a role in the development of normal prostates, but it still is unclear whether this type of AREs is also involved in the etiology or evolution of prostate cancer.
2.2.2 Mechanism of Selectivity
As discussed above, the second zinc finger plays an essential role in the binding of AR and the non-binding of GR to selective AREs. This suggests that the discrimination between the direct repeat motifs and the classical palindromes is not determined by a difference in monomeric sequence recognition, but rather by a difference in dimerization of the receptor DBDs (Helsen et al. 2012b; Verrijdt et al. 2006) . Compared to that of the GR, the AR second zinc finger differs at four positions. Two of them have been demonstrated to be involved in the stronger dimerization of AR compared to GR by creating additional interactions that stabilize the AR dimer interface (Shaffer et al. 2004) , but in functional assays, swapping these residues between AR and GR DBDs had no effect on their selectivity (Verrijdt et al. 2006) . However, removal of the carboxy-terminal part of the CTE containing these two residues abolishes AR binding to selective AREs but has no effect on AR-DBD interaction with classical AREs (Schoenmakers et al. 2000) . Deletions and point mutations in the full size receptor have similar effects on DNA binding (Haelens et al. 2007; Tanner et al. 2010) . Thus, the stronger dimerization function in the AR-DBD seems to allow the AR to dimerize on response elements that are divergent from the ARE consensus, e.g. organized as direct repeats instead of the canonical inverted repeats.
A recent study addressed the role of the DBD in ensuring specificity of AR binding to chromatin in vivo . In that study, AR binding events were compared between wild type and SPARKI mice (Sahu et al. 2014) . As discussed above, SPARKI mice express an AR that still binds classical AREs but has lost high affinity for selective AREs. Motif search analysis indicated that, as expected, an inverted palindromic repeat resembling the ARE consensus is highly enriched among the AR binding sites preferred by SPARKI AR, with a highly conserved threonine at position 12 (Sahu et al. 2014). On the other hand, analysis of the binding sites preferred by wild type AR uncovered a different element in which the 5’ hexamer is almost identical to the 5′-AGAACA-3′ consensus but followed by a second hexamer with weak sequence conservation (Sahu et al. 2014). Interestingly, the very highly conserved thymidine at position 12 present at the SPARKI AR-preferred sites is dispensable among the elements for wild type AR binding (Sahu et al. 2014). These data indicate that AR selectivity is achieved by relaxed rather than increased element stringency at chromatin binding sites which, in turn, attenuates binding of other steroid receptors to these elements. In line with this, genome-wide ChIP-seq experiments on several cell lines and tissues have shown that response elements for GR contain a highly conserved thymidine at position 12 (Grontved et al. 2013; John et al. 2011) . Altogether, these findings support the abovementioned model that the stronger dimerization of AR-DBD allows binding of a second AR molecule on 3′ hexamers that show less resemblance with the 5′-AGAACA-3′ consensus, in contrast to the GR. Interestingly, the PR also seems to have more relaxed DNA binding selectivity compared to GR (Denayer et al. 2010) .
2.3 Negative AREs
While many insights have been obtained into how the AR mediates gene activation, the mechanisms underlying AR-mediated gene repression are less understood. Possible actions include downregulation of transcription activators, competition with cofactors, and inhibition of nuclear localization of transcription factors (Grosse et al. 2012) . Another way of AR-mediated gene repression may occur through the direct binding of the AR onto the DNA and subsequent formation of corepressor complexes leading to a more condensed, thus repressive, chromatin conformation. These putative AR binding sites in the regulatory regions of androgen-repressed genes were called negative AREs.
Several examples of negative AREs have been described (Lanzino et al. 2010; Qi et al. 2012; Zhang et al. 1997) . In these papers, 5′-promotor-deletion mutants were used in reporter gene assays to identify negative AREs within the proximal region of the promotor of the androgen-repressed gene of interest. Furthermore, EMSA experiments indicated a direct interaction of the AR with the suggested ARE in vitro. Concerning the consensus sequence of these negative AREs, some authors conclude that they exhibit characteristics of partial direct repeats , thus more resembling selective AREs (Lanzino et al. 2010) . Others only see similarity with the 5′-AGAACA-3′ consensus in the 5′ part of the negative ARE, whereas the 3′ part does not resemble the hexameric consensus sequence.
While there is no doubt about the repressive effect of androgens on the expression of certain genes, incertitude remains concerning the exact nature of this transcriptional repression. Indeed, while AR binding and transrepression has been showed extensively in vitro, recruitment of androgen-activated AR to these negative AREs in a native chromatin context, e.g. by ChIP analysis, has not been demonstrated yet. Moreover, other transcription factors may participate in negative ARE function by interacting with the 3’ half of the response element. This hypothesis is supported by the presence of binding sites for Sp1 (Lanzino et al. 2010) and Ets (Zhang et al. 1997) transcription factors downstream of the described negative AREs. Thus, although androgen-mediated gene repression may involve direct AR binding to specific DNA elements, the nature of these motifs and the mechanisms by which they repress transcription remain elusive.
2.4 AREs are Always Part of Complex Enhancers
Traditionally, experimental searches for AREs have focused on relative short genomic segments just up- and downstream of the transcription start site (Claessens et al. 1989; Cleutjens et al. 1996) . In search of a more comprehensive view of androgen signaling, ChIP techniques coupled with genome-wide DNA microarray hybridization (ChIP-on-chip) or massive parallel sequencing (ChIP-seq) have identified the patterns of AR binding in various cell line models (Jia et al. 2008; Takayama et al. 2007; Wang et al. 2007) . Rather than identifying a list of target genes whose proximal regulatory elements are occupied by the AR, these studies uncovered a complex mechanism typified by AR binding to distal enhancer elements (> 10 kb). Subsequent elucidation of the mechanism by which the AR regulates transcription from afar revealed a close spatial localization of AR-bound enhancers with the promotor regions of androgen-responsive genes, which was called chromatin looping (Taslim et al. 2012; Wang et al. 2005) . Hence, AREs are not necessarily in close vicinity of the gene they regulate.
A study that combined RNA expression profiling to identify androgen-responsive genes with ChIP-on-chip to define AR binding regions (Bolton et al. 2007) demonstrated that most androgen-responsive genes are associated with two or more AREs. Moreover, the androgen-responsive genes were sometimes themselves linked in gene clusters, consisting of several androgen target genes and multiple AREs (Bolton et al. 2007). These data suggest combinatorial regulation of individual androgen-responsive genes, adding complexity to the understanding of the androgen regulation of a particular gene. That same study also confirmed the older notion that AREs appear to be composite elements, containing AR binding sequences adjacent to binding motifs for other transcriptional factors (Bolton et al. 2007; Claessens et al. 2001) . This finding was strengthened by recent AR ChIP-seq data showing that, in some cases, the AR seems to bind DNA elements as a heterodimer with FoxA1 (Sahu et al. 2011) .
Thus, androgen regulation of a target gene is more complex than AR binding on a single ARE in the promotor of that gene (Fig. 2). Indeed, AREs can also be found in distal regions, several AREs can cooperate to regulate an individual gene, and the AR may also bind as a heterodimer on a composite binding site together with another transcription factor. Additionally, as several studies report the presence of binding sites for other transcription factors in close vicinity of AREs, the AR may also bind as a homodimer on its ARE while the flanking transcription factor, e.g. GATA2, functions as a pioneer factor for AR binding (Clinckemalie et al. 2013; Wang et al. 2007) .
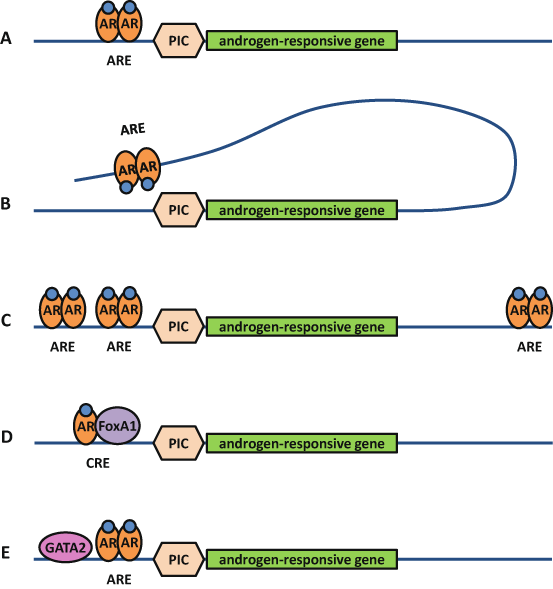

Fig. 2
Androgen response elements are part of complex enhancers. Androgen regulation of a target gene is more complex than androgen receptor (AR) binding on a single androgen response element (ARE) in the promotor of that gene a. Indeed, AREs can also be found in distal regions and make contact with the transcription preinitiation complex (PIC) by chromatin looping b, several AREs can cooperate to regulate an individual gene c, and the AR may also bind as a heterodimer on a composite response element (CRE) together with another transcription factor such as FoxA1 d. Additionally, the AR may also bind as a homodimer on its ARE while a flanking transcription factor such as GATA2 functions as a pioneer factor for AR binding e
3 Androgen Response Elements as Allosteric Modulators
While the cognate ligands of the AR are testosterone and dihydrotestosterone, the DNA response elements can also be considered ligands, rather than merely AR docking sites near the androgen target genes. Indeed, there are several lines of evidence that indicate that the response elements can modulate the activity of the bound AR, thereby acting acting as allosteric modulators.
3.1 DNA Binding Imparts Conformational Changes
Following DNA recognition, conformational changes take place in the second zinc finger that favor receptor dimerization (Freedman 1992) . As discussed above, multiple AREs are often found in the regulatory regions of androgen-responsive genes. The study of their respective role has led to the concept of cooperativity due to allosterically-induced receptor changes (Reid et al. 2001; Scheller et al. 1998) . Indeed, studies with the Slp gene enhancer show that increased AR activity is due to the interplay of several weak AREs, and that N/C-interactions contribute to this effect (Scheller et al. 1998). Cooperative interactions of multiple AREs have also been reported in the Prostate-Specific Antigen (PSA) and probasin gene promotors (Reid et al. 2001). The demonstration of cooperativity between response elements suggests the existence of binding proteins able to recognize synergy control motifs and to bridge receptor dimers bound to adjacent DNA elements. A specific amino acid motif has been postulated to be responsible for the synergy control mechanism in the GR (Iniguez-Lluhi et al. 2000) . Later on, they were identified as sumoylation sites. For the AR, the sumoylation sites in the AR-NTD have been proposed as synergy control motifs (Callewaert et al. 2004; Poukka et al. 2000) , but the exact nature of the bridging factor is still unknown. How exactly the sumoylation controls synergism is unclear, but for the AR it seems less pronounced on selective AREs (Callewaert et al. 2004), indicating a role of DNA binding.
3.2 DBD-LBD Communication
Many AR mutations have been found in patients with complete or partial androgen insensitivity as well as in biopsies of castration-resistant metastatic prostate cancer (Tilley et al. 1996) . Most of these mutations affect the function of the domain they are situated in. However, some DBD mutations do not affect DNA binding and some LBD mutations do not affect ligand binding. Surprisingly, a DBD mutation can affect ligand binding and, vice versa, an LBD mutation can affect DNA binding (Helsen et al. 2012a) . The mutated residues are situated at the surface of these domains pointing away from the DNA or the ligand. Based on modeling of the AR domains on the DBD-LBD coordinates of the crystal structure of the PPARγ-RXRα, as well as on docking AR-DBD against AR-LBD, a functional interface has been proposed between these domains, allowing signals from the DNA to reach the LBD and signals from the ligand to reach the DBD (Helsen et al. 2012a).
3.3 DBD-NTD Communication
The NTD is structurally important for AR-dependent gene expression by mediating multiple protein-protein interactions (McEwan 2004) . In contrast to the LBD and the DBD, the NTD has little stable secondary structure (Reid et al. 2002) . However, in vitro experiments with a purified AR-NTD-DBD polypeptide show that binding to an ARE results in changes in the intrinsic fluorescence emission spectrum of the NTD, indicating a conformational change in this domain upon DNA binding (Brodie et al. 2005) . Moreover, protease-resistance of the polypeptide is increased in the presence of the DNA element, adding evidence for a conformational change in the NTD upon binding to the ARE (Brodie et al. 2005). Taken together, these findings indicate that DNA binding results in an allosterically-induced conformational change within the NTD, suggesting a communication between the DBD and the NTD.
3.4 Differential Effect of Selective Versus Classical AREs
Several features of the AR have been studied by monitoring the effect of point mutations on the functionality of the receptor in reporter assays. The effects of disrupting the N/C interaction, sumoylation of the NTD and the role of the polyglutamine stretch in the control of the overall activity of the human AR have initially all been described on reporter genes controlled by classical AREs (Claessens et al. 2008) . However, the same analyses performed with reporter genes based on selective AREs gave much less pronounced or no effects. Indeed, deletion of the FQNLF motif in the NTD, which blunts the N/C-interaction, strongly reduced the AR capacity to transactivate through classical AREs but did not affect its activity on selective elements (Callewaert et al. 2003b) . Mutation of lysine 385 in the NTD, blocking sumoylation at that site, clearly affects the cooperativity of the receptor on multiple classical AREs. However, when selective AREs where tested, the lysine 385 mutation did not increase the androgen response (Callewaert et al. 2004). Deletion of the polyglutamine tract results in an increase in the transactivation through classical AREs, whereas this effect is not observed on selective elements (Callewaert et al. 2003a). Finally, modulation of AR activity by coactivators, AR sensitivity to proteolysis, and the effect of mutations in the AR dimerization interface of the DBD also vary with the nature of the ARE, the observed effect being different when using classical AREs compared to selective ones (Geserick et al. 2003) .
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree