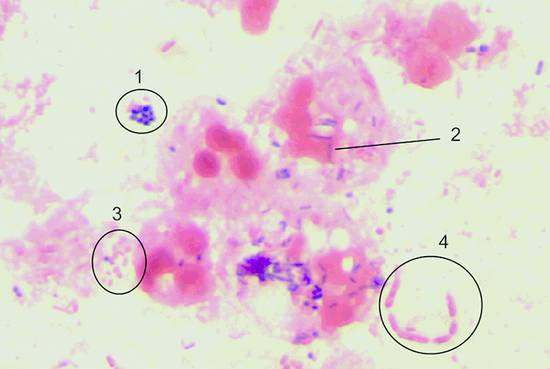
Infections and Diagnostic Microbiology Specimens for microbiological testing can be collected from numerous organs and tissues by an experienced examiner using ultrasound-guided aspiration and biopsy: Ascites Cyst fluid Abscess contents Pleural effusion Pericardial effusion Synovial fluid Liver tissue Renal tissue Lymph node tissue When a specimen is collected by percutaneous needle aspiration, rigorous precautions should be taken to avoid transferring pathogens from the skin surface to deeper tissues and to ensure that the sample is not contaminated by skin flora. Skin preparation is essential, therefore. The antiseptic solutions most commonly used for this purpose are 10% PVP-iodine and alcohols. Alcohol-based solutions appear to be more effective than aqueous iodine solutions and mixtures of alcohols and iodine or chlorhexidine.1,2 Skin preparation should include mechanical cleansing of the site by scrubbing with a sterile gauze pad. Hair-bearing skin should not be shaved before the puncture, since shaving causes tiny skin abrasions that predispose to local infection. Any hairs at the puncture site may be carefully clipped with a scissors, however. Recommendations on necessary exposure times to antiseptic solutions must be observed. Most experts recommend an exposure time of 60 to 180 seconds. However, skin areas rich in sebaceous glands (e.g., the groin) may require a longer exposure (up to 10 minutes) to achieve an adequate reduction of microorganisms. It is good practice to collect as much material as possible so that there will be enough for chemical, cytological, and microbiological tests. Scanty specimen volumes can reduce sensitivity and increase sampling errors. ▶ Table 7.1 lists recommendations on minimum specimen volumes required for different diagnostic tests. These figures may vary somewhat from laboratory to laboratory, depending on the specific methods used. Communication with the microbiologist is advised, especially with scanty specimens, so that the best culture method can be used. If tissue for pathologic examination is collected in addition to material for microbiological testing, meticulous care should be taken that the sample for microbiological testing does not come into contact with formalin solution, as that would preclude any further microbiologic analysis. Before a specimen is sent for microbiological testing, it needs to be placed in a transport container that is appropriate for the specimen and the desired tests. The type of container depends on the expected transport time and presumptive organisms. If transport conditions are unfavorable, the culture result may be tainted by the destruction of delicate microbes and the overgrowth of more robust organisms. If the specimen can be transported to the microbiology laboratory in less than 2 hours, it is generally sufficient to submit a fresh specimen in a sterile tube (e.g., in the aspiration syringe itself). If transport time will exceed 2 hours, even with immediate submittal, a suitable transport medium must be used. Blood culture media are excellent for liquid specimens. In this case, however, additional fresh material should always be submitted so that microscopic preparations, possible antigen tests, cultures for mycobacteria or fungi, and other special cultures can be grown. If the specimen were inoculated only into blood culture bottles, none of these tests could be performed; nor could a Gram stain be done to provide a plausibility check for culture results. Another option is to use a semisolid, prereduced transport medium (e.g., Port-A-Cul system, or similar). Microbiology laboratories will usually provide necessary submission supplies at no charge (▶ Fig. 7.1). Fig. 7.1 Submission containers. a Submission container for glass slides (bottom); transport container with semisolid prereduced transport medium (e.g., Port-A-Cul [Becton Dickinson]) (center); and swab kit with gel chamber (top). b Containers for submitting liquid specimens (from left to right): two anaerobic/aerobic blood culture bottles, transport container with semisolid prereduced medium (e.g., Port-A-Cul), sterile tube, syringe with stopper. If there is clinical evidence that the lesion of interest has already given rise to a systemic infection, blood culture bottles (at least two pairs of aerobic and anaerobic) should also be submitted. With modern blood culture systems, care should be taken that the aerobic blood culture bottle is not “aerated” (risk of contamination). These systems provide optimum aerobic culture conditions with no need for additional measures. Transport to the microbiology laboratory should be as rapid as possible. If collected specimens cannot be submitted right away, blood cultures should be incubated at 35°C and all other materials stored at 4°C to prevent specimen contamination or overgrowth by fast-growing organisms. Specimens must be labeled with the name of the patient, the sender, and specimen information and submitted to the laboratory with the proper request form. Information on the nature of the specimen along with clinical information and the presumed pathogen are essential so that the microbiologist can select the most appropriate culture techniques and detection methods. Macroscopic examination of the specimen should be done immediately after collection. If facilities are available, a portion of the specimen can be microscopically examined on-site by Gram staining a fresh sample. Gram staining (see below) can provide fast, valuable information on the causative organism(s), especially if the material cannot be sent to the microbiology laboratory right away. Staining is the first step in microbiological testing. Stains can be performed in the microbiology laboratory immediately after specimen receipt or even on the ward and can provide an early source of valuable information. Stains supplement culture methods and often aid in their interpretation. This particularly applies to Gram staining. In addition, microscopic examination permits a semiquantitative assessment of the pathogen distribution in the specimen and an estimation of the degree of inflammatory response (WBC count in the specimen). Parasites (amebas, echinococci, etc.) can also be detected microscopically. The Gram stain is the most important stain in microbiology. It differentiates between gram-positive and gram-negative organisms owing to the fundamentally different cell wall structures of these two types of bacteria. Another advantage of the Gram stain is that it can be done quickly, even outside the microbiology laboratory. The supplies necessary for Gram staining are illustrated in ▶ Fig. 7.2 and include the following: Fig. 7.2 Workstation with supplies for Gram staining. A microscope (×10, ×40, ×100/oil) Glass slides Bunsen or gas burner Acetone or methyl alcohol Crystal violet 0.25% Lugol solution 1% (iodine and potassium iodide) 96% ethyl alcohol Safranin (fuchsin) 0.1% For an outline of the Gram-staining procedure refer to ▶ Table 7.2 and ▶ Fig. 7.3. A Gram-stained preparation is illustrated in ▶ Fig. 7.4. Fig. 7.3 Schematic representation of the Gram staining process. Fig. 7.4 Gram stain of abscess material with polymorphonuclear leukocytes in various stages of degeneration, gram-positive cocci (1), gram-positive rods (2), gram-negative cocci (3), and gram-negative rods (4) (×1000 magnification). The most important gram-positive and gram-negative pathogens are listed in ▶ Table 7.3. Ziehl–Neelsen and auramine stains are used almost exclusively in microbiology laboratories for the detection of acid-fast rods (mycobacteria, Nocardia spp., Actinomyces spp.). Since bacterial counts in specimens are usually low, fluorescent stains (auramine stain, ▶ Fig. 7.5) are often used to increase the sensitivity of microbial detection (see the Box ▶ “Key questions relating to the differential diagnosis of enlarged lymph nodes”). Fig. 7.5 Acid-fast rods in abscess material visualized by auramine staining Other fluorescent stains are available in the laboratory for the microscopic detection of microorganisms. These stains can reveal fungi or bacteria that are difficult to stain by other methods. Amebas and other parasites are on very rare occasions found in aspirates and are easily confused with leukocytes. It is important to inform the microbiologist of their suspected presence. On the other hand, serology is an important adjunct in patients with suspected parasitic diseases such as amebic liver abscess or hydatid liver disease, especially because needle aspiration of hydatid liver cysts should be avoided, as this could cause peritoneal seeding of worm larvae (protoscolices). Infections by Echinococcus spp. are detectable by serologic testing in almost all cases; only well-encapsulated cysts of Echinococcus granulosus (dog tapeworm) may be serologically negative in rare cases. The isolation of microorganisms from invasively sampled materials is done under varying growth conditions on a combination of liquid and solid nutrient media. With fast-growing organisms, a preliminary differentiation can be made in just 24 hours. By 48 hours, it is generally possible to make a definitive identification and perform susceptibility testing in a bacterial isolate. In modern microbiology laboratories, this is usually done in fully automated incubators using standardized culture strips. Many specific and nonspecific nucleic acid amplification techniques are currently available to detect the DNA of microorganisms. Further differentiation is possible by sequencing of the amplification products. These techniques are particularly useful for detecting pathogens that are slow-growing or difficult to culture, such as viruses (DNA, RNA), and for the detection of pathogens in patients on antimicrobial therapy. Mycobacterial DNA is still detectable in tissue for years after the complete resolution of tuberculosis. Specific assays are available for the Mycobacterium tuberculosis complex, for atypical mycobacteria, for Aspergillus, for Rochalimaea, for toxoplasmosis, and for many other pathogens, in specialized microbiology laboratories. Bacterial 16S ribosomal DNA shows variable and highly conserved regions. Using primers that recognize the conserved regions, the variable sequences located beneath the highly conserved regions can be amplified, sequenced, compared with entries in gene banks, and then assigned to a particular species. The 16S rDNA assay is generally a highly sensitive procedure, but it may be compromised by nonspecifically inhibiting substances in the sample. The test is effective only for materials that are sterile under normal conditions but is still susceptible to contamination due to its high sensitivity. To date, nucleic acid amplification techniques for genotypic resistance analysis have been developed only for certain resistance mechanisms and only for a few organisms (e.g., MRSA). They have so far been unable to replace time-consuming culture methods. Serologic tests are of limited value for the detection of acute infections because a detectable antibody response appears only after a latent period of several days. Ordering serologic tests makes sense only if the result will have diagnostic or therapeutic implications. The confident serologic diagnosis of an infection generally requires drawing a second sample 1 to 2 weeks later that will show a significant rise in antibody titers. There are only a few serologies for which a single high IgG or IgM titer is diagnostic of an infection. Within the context of this book, these diseases include echinococcosis, amebiasis, schistosomiasis, hepatitis B/C, and HIV infection. Serologic analysis requires experience. Physicians should resist the indiscriminate ordering of serologic tests that are not appropriate for the disease in question. Another pitfall is the overinterpretation of positive IgM titers, especially if the titers are low. This is usually a nonspecific finding. The turnaround time for issuing a final report on a microbiology sample is usually 48 hours for fast-growing organisms. Occasionally, however, even preliminary findings can help to optimize treatment and can be obtained simply by calling the laboratory. Average turnaround times are listed in ▶ Table 7.4. Exact times will vary depending on sample throughputs and laboratory organization. The accuracy of microbiological testing depends directly on the desired scope of the tests before the sample is submitted. It should be emphasized that Mycobacterium tuberculosis, for example, would be missed in a sample submitted for testing for “pyogenic organisms” or the “presence of organisms” even if the sample contained massive numbers of the mycobacteria. It is essential, therefore, to request tests that are consistent with clinical information. A specific request is also helpful for limiting costs. In equivocal cases, it is advantageous to discuss the scope of the tests with the microbiologist or the infectious disease specialist. Moreover, the test results will depend in large measure on the quality of the material that is collected and submitted for analysis. Common reasons for unsatisfactory microbiological test results include the following. Sampling error due to an inadequate specimen volume Sampling error due to low bacterial counts Slow or delayed transport, improper transport container, failure to culture fastidious pathogens (e.g., anaerobes) due to overgrowth of other organisms Detection of ubiquitous organisms, typical (skin) commensals or contamination (e.g., coagulase-negative staphylococci, corynebacteria, etc.), especially when determined in liquid culture media, blood cultures or PCR assays, since only a few organisms are sufficient to produce a positive result in these procedures. The significance of detecting these organisms must be questioned, especially when they do not match the clinical information and/or the Gram stain result. A Gram stain that matches the isolated organism, or the repeated detection of one of these organisms, is less suggestive of contamination and is more consistent with an infecting organism. Previous antimicrobial therapy In purulent materials (i.e., materials containing large numbers of white cells), the antimicrobial substances released from degenerating granulocytes may produce false-negative culture results. Consequently, purulent materials should always be diluted in transport media (e.g. in blood culture bottles) when submitted for testing. Inadequate request or unreasonable expectation for microbiological testing, such as the growth of mycobacteria in ordinary blood culture bottles, the growth of parasites in a bacterial culture, and so on Expecting culture results within a few hours Nonculturable organisms (e.g., parasites)
7.1 General Principles of Microbiological Testing
7.1.1 Microbiological Specimens
7.1.2 Prerequisites for Microbiological Testing
Skin Preparation
Specimen Collection and Volume
Test
Absolute minimum volume
Ideal volume
Gram stain
0.2 mL
0.5 mL
Aerobic culture
0.2 mL
2 mL
Anaerobic culture
0.2 mL
2 mL
Fungal culture
0.2 mL
2 mL
Mycobacteria culture
3 mL
10 mL
Blood culturesa
2–10 mL
10 mL
Borrelia burgdorferi—PCR
2 mL
5 mL
PCR testing
0.2 mL
1 mL
Serology (blood)
3 mL
10 mL
aIt is important to follow manufacturer’s recommendations.
Specimen Submission to the Microbiology Laboratory

Immediate Examination of Specimens
7.2 Microbiological Techniques
7.2.1 Stains
Gram Stain

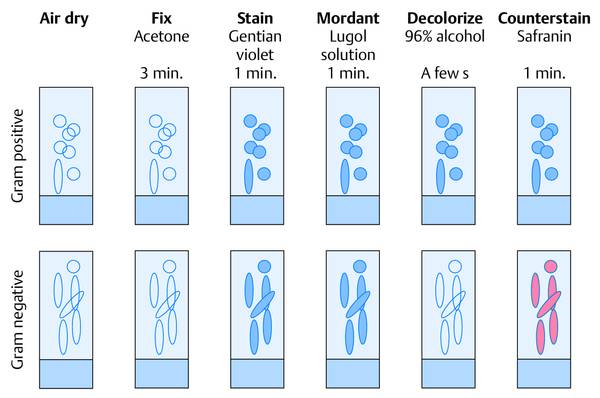
Step in procedure
Action
1
Label the frosted end of the slide with a pencil.
2
Smear out the aspirated material (may dilute with sterile saline if necessary).
3
Air-dry the smear.
4
Fix: cover with acetone or concentrated methyl alcohol for 3 min or pass 3 times through the burner flame.
5
Cover with crystal violet for 1 min, pour off.
6
Cover with Lugol solution (iodine–potassium iodide), pour off.
7
Decolorize with 96% alcohol (until all blue is gone).
8
Rinse with water.
9
Cover with safranin.
10
Rinse off with water and dry.
Tips
Adequate decolorization is essential, so a thin smear is desired.
Check: all body cells should stain red; blue staining means inadequate decolorization.
The lower limit of detection is approximately 104 organisms/mL.
Gram-positive cocci
Gram-positive rods
Gram-negative cocci
Gram-positive rods
Staphylococci
Clostridia
Neisseria spp.
Enterobacteriaceae
Enterococci
Cornybacteria
Acinetobacter spp.
Pseudomonas spp.
Streptococci
Bacillus spp.
Bacteriodes spp.
Ziehl–Neelsen Stain, Auramine Stain
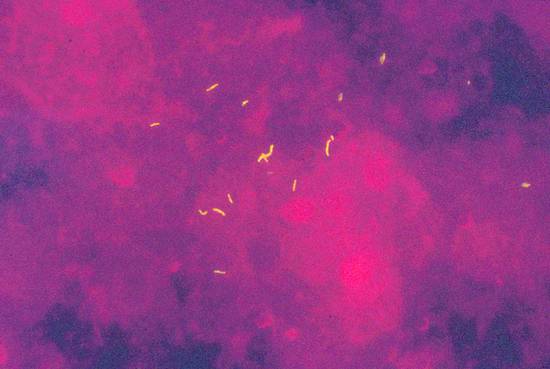
Detection of Parasites
7.2.2 Culture Techniques
7.2.3 Nucleic Acid Amplification Techniques
7.2.4 Serology
7.2.5 When Are Microbiological Test Results Available?
Result within:
Tests
1–3 hours
Gram stain, antigen detection, specific PCR (e.g., LightCycler [Roche Applied Science])
3–5 hours
Detection of acid-fast rods, other special stains
24 hours
Preliminary differentiation of fast-growing bacteria.
Result of antimicrobial susceptibility testing done directly from patient samples (e.g., positive microscopic findings).
Growth in blood cultures (yes/no, Gram stain)
48 hours
Differentiation and susceptibility testing of fast-growing aerobic organisms
>72 hours
Differentiation of anaerobes
Approximately 14 days
Mycobacteria in liquid culture, rapidly growing atypical mycobacteria (MOTT, NTMa)
4–6 weeks
Mycobacteria in solid culture, slow-growing organisms
aMOTT, mycobacteria other than tuberculosis. NTM, nontuberculous mycobacteria.
7.2.6 Limitations of Microbiological Methods
7.2.7 Specimen Receipt during Off-hours (Nights, Weekends, Holidays)
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree