Fig. 1.1
Preoperative 1.5T (left), intraoperative 0.5T (middle), and combined (right) T2W MR images. In the combined image, registered preoperative imaging is shown within the prostate, while in the surrounding area, the intraoperative image is shown. Note that significant soft tissue deformation has occurred between scans (Haker et al. [4])
Multimodality and Multiparametric Imaging
The choice of the appropriate imaging modality and the selection of the right imaging parameter are essential for satisfying the demands of a complex IGT procedure. All modalities represent different physical probes that reflect different physical properties of the imaged tissues. In medical images the information is composed from spatially localized measurements that result in spatially distributed intensities that represent x-ray absorption, acoustic pressure in US, and RF signal amplitude in MRI. All imaging modalities have limitations and advantages, and all have unique and characteristic artifacts. Combining multiple modalities by image fusion is a powerful tool to emphasize advantages and eliminate disadvantages of each applied individual modalities. X-ray and US are bona fide real-time imaging modalities. However, advanced cross-sectional imaging devices like CT and MRI can provide near real-time “fluoroscopic” imaging with sufficient temporal resolution for most IGT procedures. CT demonstrates anatomy, and PET metabolic or functional data, but their integration in PET/CT provides a full definition of both the lesion and the related anatomy. Even more advantages of integrated multimodality imaging are evident when PET is combined with multiparametric MRI. Figure 1.2 demonstrates image fusions.
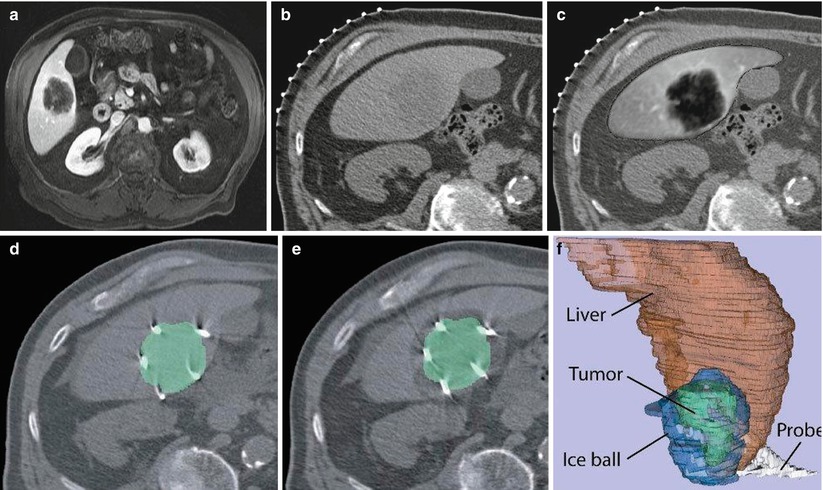

Fig. 1.2
Image fusion between CT and MRI. (a) Preprocedure contrast-enhanced MR image, (b) CT image for interventional planning, (c) registered and fused image showing liver from MR image overlaid on CT image, (d) CT image showing ablation probes overlaid with segmented tumor from the registered MR image, (e) CT image showing iceball (hypointense area around lesion) overlaid with segmented lesion (green) from registered MR image, and (f) 3D representation of liver (red), tumor (green), iceball (blue), and cryoprobes to evaluate tumor coverage and margins
Most of the imaging modalities have multiple parameters to choose from for a particular imaging task. Multiparametric imaging enhances tissue characterization and provides a preliminary feature extraction before image processing. For example, x-ray can demonstrate bones and, after contrast injection, vascular structures; CT can be displayed, showing only bones, vascular anatomy, or tissue perfusion; US can detect soft tissues and depict blood flow (Doppler mode) but also can represent various tissue parameters (elasticity, stiffness). MRI is the ultimate multiparametric imaging modality because the radiologist can select from large numbers of relaxation, diffusion, and dynamic contrast-enhancement (DCE) parameters to improve the sensitivity and specificity of a given imaging task [5, 6]. In addition, several imaging methods are available to separate fat from water, cartilage from bone, and to visualize vascular structures without contrast administration. MRI contrast mechanisms depend on a number of tissue parameters, and many different pulse sequences are available to emphasize different tissue characteristics. Choosing the optimal pulse sequence influences not only image quality but also image processing, especially segmentation. It requires knowledge of the underlying tissue properties of the anatomy to be segmented. Imaging artifacts and noise, partial volume effects, and organ motion can also have significant consequences on the appearance of images and the performance of segmentation algorithms. Improving image quality does not necessarily improve segmentation. Increased resolution may result in errors in image analysis methods.
Image Processing
It is well known that humans can distinguish different image regions or separating different classes of objects and can perform such tasks with ease. Computers are not able to distinguish anatomical structures, lesions, or any other essential features without processing the information. The goal of medical image processing is to extract quantitative information from various medical imaging modalities to better diagnose, localize, monitor, and treat medical disorders.
Segmentation
Segmentation is defined as the partition of an image into overlapping regions that are homogeneous with respect to image intensity or texture. The purpose of segmentation is to provide richer information than that which exists in the original medical images alone. Segmentation identifies or labels any structures that are represented by signal intensities, textures, lines, or shapes. The resulting segmented features can define volumes, shapes, and locations and/or provide visualization or quantization by surface or volume rendering.
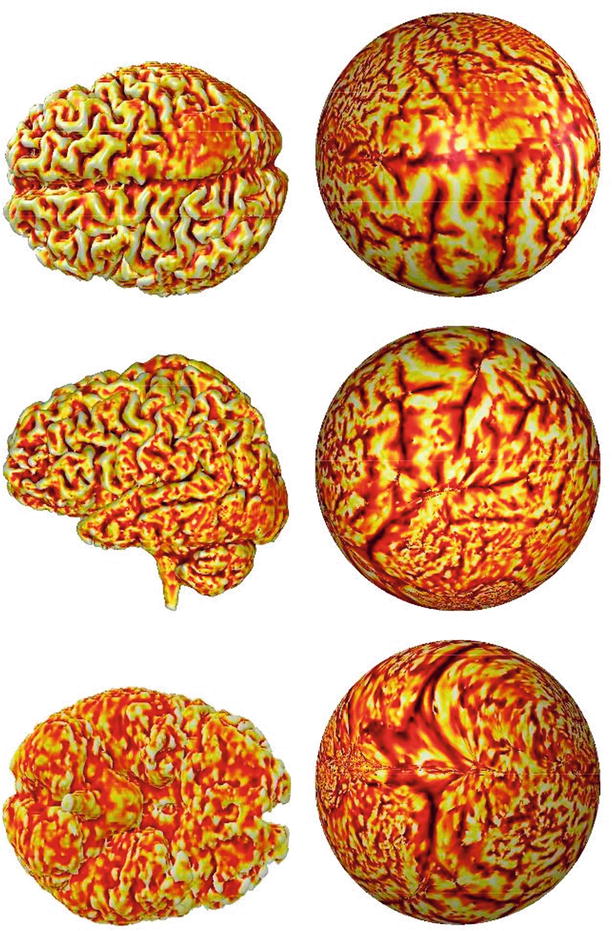
During segmentation, a label is assigned to every image component (in 2D digital images to pixels and in 3D to voxels) that shares certain characteristics. Image segmentation is typically used to locate objects and boundaries (lines, curves, etc.) in images. Segmentation methods are based on signal intensity, texture, edges, and shapes. The segmentation method can operate in a 2D or a 3D image domain. Usually, 2D methods are applied to 2D images, and 3D methods are applied to 3D images. In some cases, however, 2D methods are applied to 3D multislice images, a change that requires less computation as some structures are more easily defined on 2D slices. More difficult is medical image segmentation on a surface that is a 2D object folded in a 3D space like the cortex of the brain. This type of segmentation is not a simple 2D or 3D problem though it is possible to flatten a surface into a 2D plane in a conformal manner [7, 8]. Figure 1.3 shows three views of flattened brain surface.
Segmentation can also be thought of as the division of an image into clinically relevant components. It can be done with operator control, automated signal intensity, or template (anatomical knowledge)-driven techniques. The most straightforward segmentation method is operator-controlled segmentation in which an expert is used to guide the process. It is tedious and prone to error, especially if the structure of interest does not align well with the planes of the image acquisition. An alternative approach is to use properties of signal intensity to drive the segmentation. In the case of MRI, inhomogeneous signal intensities are problematic. Intensity-based classification of MR images has proven challenging, even when advanced techniques are used. Intrascan and interscan intensity inhomogeneities are a common source of difficulty. While reported methods have had some success in correcting intrascan inhomogeneities, such methods require supervision for the individual scan. Adaptive segmentation uses knowledge of tissue intensity properties and intensity inhomogeneities to correct and segment MR images. Use of the expectation–maximization (EM) algorithm leads to a method that allows for more accurate segmentation of tissue types as well as better visualization of MRI data [9–11].
Medical image segmentation is a difficult process for two primary reasons: image acquisition is usually suboptimal for representing the tissue of interest, and the relevant anatomy is too complex to optimally plan and guide surgery. In US, MR, CT, and PET, in most cases, the tissues of interest are inhomogeneous or not clearly separable from the surroundings. Their grayscale value is not constant, and strong edges may not be present around borders. As a result, on the images the regions that are detectable by the human eye cannot be segmented even with sophisticated computer algorithms. Image segmentation algorithms may not work well in the presence of poor image contrast, significant noise, and smeared tissue boundaries (due to organ or patient movements). In this case, the inclusion of as much prior information as possible helps the segmentation algorithms to extract the tissue of interest. The complexity and variability of the anatomy make it difficult to locate or delineate certain structures without detailed anatomical knowledge. Shape- and atlas-based approaches are helpful to improve segmentation [10, 12, 13]. Figure 1.4 shows an example of space-controlled probabilities of anatomical structures used for segmentation. Today a computerized algorithmic approach does not even come near to the expert knowledge of a trained radiologist. The knowledge must be built into the system or provided by an experienced human operator. An atlas can be generated by compiling information from a large number of individuals. This atlas is then used as a reference frame for segmenting any individual images. Semi-automated or fully automated segmentation methods are necessary for intraprocedural segmentation and for the subsequent generation of 3D models. Today, still, mostly manual and expert-guided interactive segmentation is used to enable clinicians to process images in very general circumstances but in a very time-consuming way. New methods for patient-specific image segmentation are needed to take into consideration the heterogeneity and variability of anatomy and pathology.

Fig. 1.4
Examples of space-controlled probabilities of anatomical structures. Blue indicates low and red high probability in respect to a presence of a structure at that voxel location (Pohl et al. [12])
The use of image segmentation is essential in IGT applications, especially in clinical settings. To remove tumors or to perform biopsies or thermal ablations, physicians must follow complex multiple trajectories to avoid critical anatomical structures like blood vessels or eloquent functional brain areas. Before surgery, path planning and visualization is done using preoperative MR and/or CT scans along with 3D surface models of the patient’s anatomy. During the procedure, the results of the preoperative segmentation may still be used if the surgeon has access to the preoperative planning information as 3D models and grayscale data are displayed in the operating room. In addition, “on-the-fly” segmentation of real-time images generated during interventions and/or surgeries have been used for quantitative monitoring of the progression of surgery in tumor resection and ablation [14].
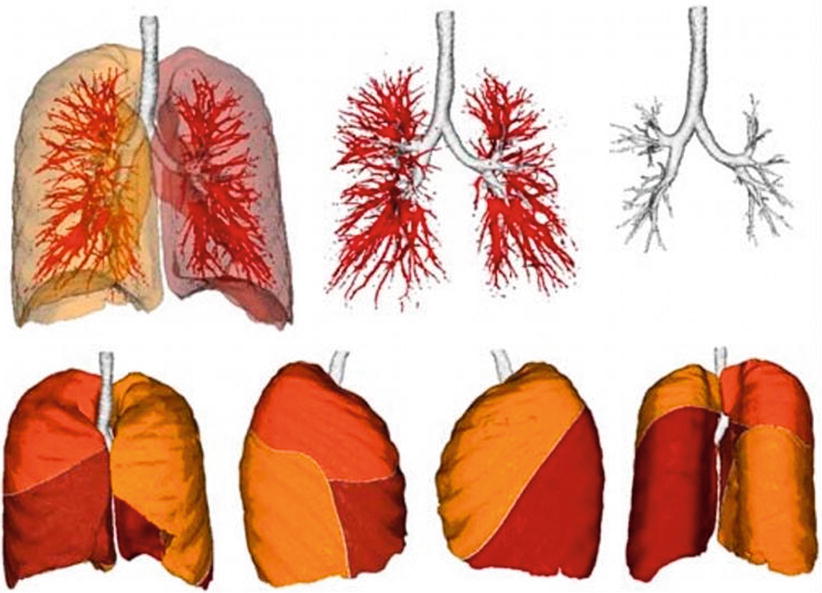
When evaluating segmentation methods for a particular application, the following factors are important: running time, the amount of user interaction, accuracy, reproducibility, and applicability to the clinical situation. Despite much research effort [15–17], a fully automatic solution to the long-standing image segmentation problem remains unattainable. Figure 1.5 shows fully automatic segmentation of the lung (lobes) and airways.
Registration and Image Fusion
In IGT, the real patient and the image-based model (virtual patient) have to be co-registered in the same reference frame. Registration is the transformation of images and the patient’s anatomy into this common reference frame. In the registration process, we spatially match datasets that were taken at different times or by different imaging modalities or in different planes. If registration is correct, spatial correspondence between anatomical, functional, or metabolic information can be established.
The geometric alignment or registration of multimodality images is essential in IGT applications. The process of patient registration involves matching preoperative images to the patient’s actual coordinate system. The various registration algorithms are either frame based, landmark based, surface based, or voxel based. Stereotactic- or frame-based registration is accurate, but outside the brain is not practical and cannot be applied retrospectively. Landmark-based methods use either external point landmarks or anatomical point landmarks, and their accuracy is influenced by skin tissue and organ motion. Surface-based registration uses surface segmentation algorithms, and if the surfaces are not easily identified, they may fail. The accuracy of voxel-based registration methods is not limited by segmentation errors as in surface-based methods.
Several anatomical landmarks common to both the patient and the preoperative images have to be identified. These landmarks serve to register the images to the patient’s coordinate system during a surgery. Since human anatomy is not well defined, fixed landmarks that can be accurately identified as fiducial markers are added for registration. Fiducial markers are often stuck to the patient’s skin prior to the scan, making them tactile and also visible on images. Skin movement inevitably causes errors in registration. Affixing the fiducial markers directly to bone is used to avoid this movement. Developments in intraoperative imaging and navigation can help to abolish fiducial markers and allow for real-time or automatic registration.
Image acquisition and intraoperative image processing have improved steadily in recent years due to increasingly sophisticated multimodality image fusion and registration, although most methods are confined to rigid structures. The deformations of normal anatomy during surgery obviously require the application of nonrigid registration algorithms and updates of the anatomical changes using intraoperative imaging. Image registration is rigid when images are assumed to be solid and only need rotation and/or translation to be matched with each other. Nonrigid registration is necessary when structures present in both images cannot be matched without some localized stretching, warping, or morphing of the images. In most of the IGT applications, rigid registration is not accurate, and much of the registration today involves nonrigid registration techniques [15, 19–22].
Clinical experience with IGT of the brain and prostate and in surgeries with large resections or displacements has revealed the limitations of rigid registration and visualization approaches and resulted in the development of multiple nonrigid registration algorithms [19, 23–25]. IGT registration approaches require minimum user interaction and are compatible with the time constraints of our interventional or surgical clinical workflow. The automatic deformable registration also should have acceptable accuracy, reliability, and consistency [26].
Patient-Specific Modeling
Image processing is essentially patient-specific analysis of images with the goal to obtain multiple images from individuals with pathologies that deviate from the normal population image datasets. These patient-specific models differ significantly from one patient to another or from one time point to another and reflect the patient’s distinct personal anatomy, localized pathology, and, in some cases, functioning. Image processing tools for IGT rely on algorithmic approaches that consider substantial anatomical similarity between individuals and are based upon the geometric regularity and stability of anatomy and function. This reliability or predictability emphasizes prior knowledge but preserves individual deviations or alterations and, therefore, enables more effective interpretation of images. Various statistical models can represent a wider range of anatomies and pathologies [12]. Differences in anatomy caused by tumors that displace the surrounding structures can be handled with the same statistical model. Finding anatomical associations between patients, groups of patients, and individual patients over time is an important part of image analysis. Figure 1.6 shows a patient-specific 3D model of a renal lesion and surrounding anatomical structures.
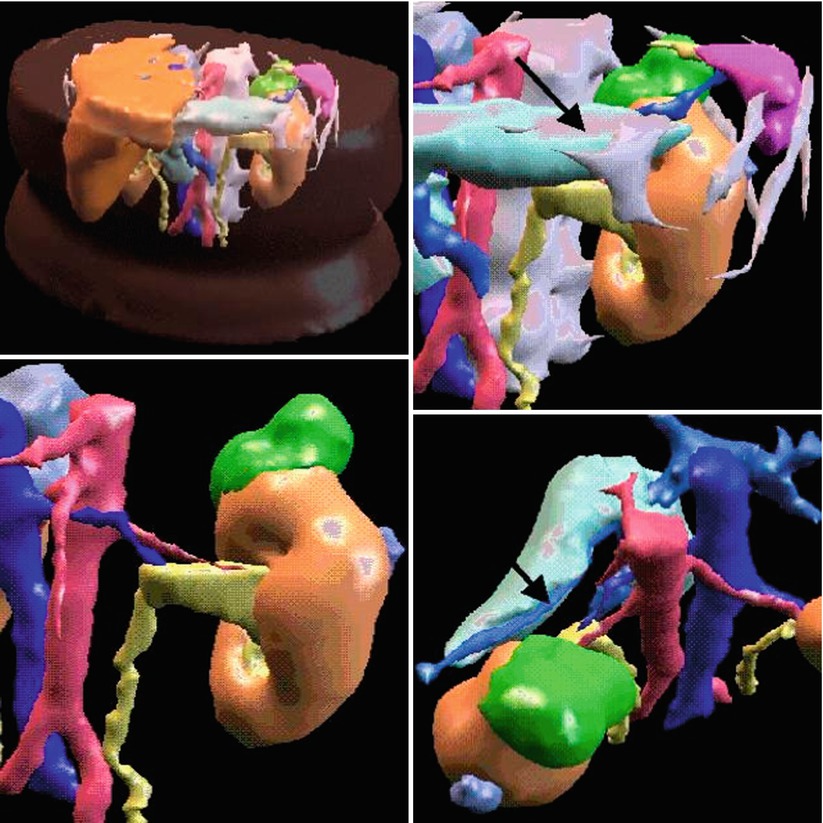

Fig. 1.6
3D model of a renal lesion and surrounding structures. Renal tumor is green, the arrow shows to the pancreas which is close within the retroperitoneal space (Young et al. [27])
Cross-sectional images have to be rendered in 3D to make a representation of the individual patient: the virtual patient. When image processing algorithms are applied to multislice images, the resulting contours after image segmentation can be used to create 3D reconstructions with the help of interpolation algorithms like marching cubes [28]. In marching cubes segmentation, isosurfaces are found along a chosen grayscale value, essentially separating voxels of a higher value from voxels of a lower value. The algorithm places cubes connecting the voxel centers, and if the isosurface lies in the cube, it decides what type of local surface polygon should pass through the cube. A limited number of possible surface topologies exist to allow this local approach to rapidly construct a 3D polygonal model. This type of segmentation is directly applicable to medical image data when the desired structure has very clear and constant boundaries, such as bone in a CT image.
In IGT the 3D model has to be a dynamic one because it may change during or after procedures. If serial or longitudinal images are collected over time (time series) of the same subject, 4D dynamic models can be generated that can be used to interpret disease progression and monitor response to therapy as they represent intraoperative dynamic changes like shifts and deformations.
Patient-specific information is in the images, but processing is necessary to reveal their representation in the model. The virtual reality of the model and the plan is registered to the actual reality of the patient to help the clinician perform the planned procedure. Given that the difference between simulation and planning is minimal, 3D models can be used for both surgical simulation and/or surgical planning. In both, similar image processing methods are used to change and manipulate the data according to the simulated or planned surgical process. The goals of simulation and planning, however, are different; simulation is used mostly for training, and planning is used to optimize the surgery. Simulation can improve planning. Both processes are used in a similar virtual environment. There are some tools that are primarily useful for simulations to interact with the virtual model, such as collision detection, compliance, haptic interfaces, tissue modeling, and tissue tool interactions. These and biomechanical models that explain tissue deformations can be applied to planning surgical procedures [23, 29]. The best example to demonstrate the overlap between surgical simulation and planning is robotic surgery. Instead of directly moving the instruments, the surgeon uses a computer console to remotely manipulate the instruments attached to robotic arms. These systems can be used both for training as robotic surgical simulators and performing real robotic surgeries on patients.
Image Display and Visualization
Advances in medical visualization are combining 3D rendering techniques with novel spatial 3D displays to provide all the available information to the human visual system. As the surgical opening becomes smaller, the visualization of the direct surgical site is becoming increasingly difficult. Virtual reality techniques can be used, and model-based surface or volume rendering and computer graphics can be applied to emphasize structures from CT or MRI to allow the surgeon to navigate and rehearse a surgical situation without directly affecting the patient. However, none of the display techniques can represent all 3D data to the user in an optimal way. Even if the displayed image looks strikingly realistic, it may not be useful for image guidance. No single graphical user interface can satisfy all users and tasks; therefore, it is necessary to form an adaptive graphical interface that integrates all imaging information and accounts for all user objectives.
If visualization requires opaque surfaces, polygon-based surface rendering techniques are used, which is a method that classifies the parts of images into objects and non-objects. This binary surface extraction is practical for objects that have sharp boundaries like skin or bone, but it is quite difficult for most soft tissues and for tumors for which boundaries are less defined and surfaces are fuzzy. For this type of tissue, another visualization technique is used: volume rendering that does not require binary classification. Both visualization techniques, however, require active user interaction by using different viewing positions, the removal of overlaying surfaces or volumes, and the highlighting of others, etc.
Segmentation is defining a structure in the reference frame of images; registration is transforming this to another image-based reference frame or to the patient’s own frame of reference. Navigation serves to localize one or more points in this integrated frame of reference. Visualization refers to seeing this from the user’s (physician) frame of reference. However, if the physician enters into the common reference frame, it will be part of an augmented reality system. In IGT the real patient and the image-based virtual patient have to be co-registered in the same reference frame. If the real and the virtual object are integrated and all navigation and interaction are in the same system, it is called an augmented reality system. In this situation, the operator is using various virtual reality interaction devices with trackers (3D gloves and/or googles, 3D head-mounted devices, etc.) to interact with the unified system. A good example is interactive virtual endoscopy with which the endoscopic operator navigates the tracked endoscope in a virtual patient-specific model [30]. Integration of real and imaginary systems plays a major role not only in IGT but also in telesurgery.
In IGT, integrated visualization and image analysis platforms like the 3D Slicer can be used [31]. The 3D Slicer (Fig. 1.7) provides a graphical user interface to interact with data. In addition to manual segmentation and the creation of 3D surface models from conventional MR images, Slicer has also been used for both rigid and nonrigid image registration [33].
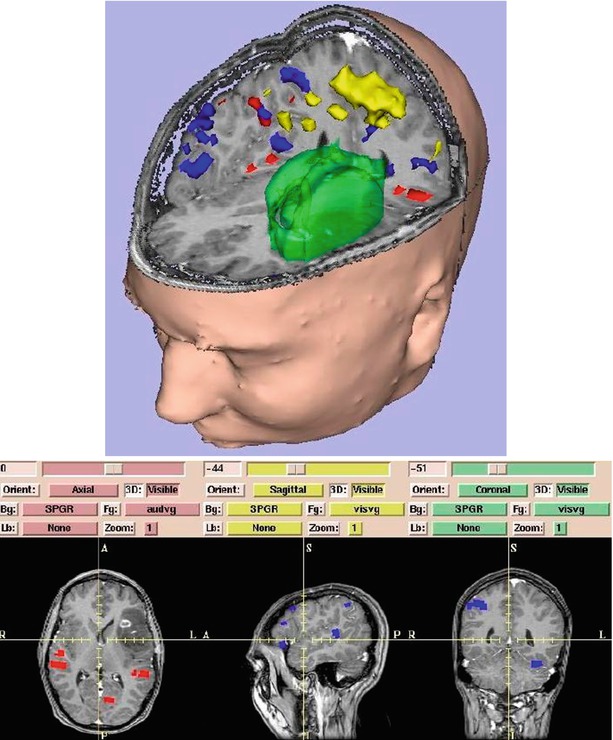

Fig. 1.7
Surface models of skin, tumor (green), motor cortex (yellow), auditory verb generation (red), and visual verb generation (blue) are integrated with slices. On 3D Slicer (Gering et al. [32])
Guidance
Images provide spatially defined information about anatomy, function, or other image extractable features that can be used for guiding procedures or devices. This human-controlled guidance requires a high degree of human interaction. Guidance systems navigate without direct or continuous human control. They consist of three essential parts: navigation that tracks current location, guidance that combines navigation data and target information, and control that accepts guidance commands to reach the target. Navigation is to find out “where you are,” guidance is to decide “where to go,” and control is to direct the system “how to go there.”
Navigation
Navigation in surgery is based upon stereotactic principles to locate a point or points in a reference frame. Image-guided navigation minimizes the guesswork associated with complicated surgery and helps surgeons to perform surgery accurately. The navigation systems can also establish a spatial relationship between preoperative images and the intraoperative anatomy. A specific point in the imaging dataset is then matched with the corresponding point in the surgical field using registration. To provide accurate data for navigation, a minimum of three points should be registered or matched. After registration is completed, the probe can be placed on any preferred point in the surgical field, and the corresponding point in the imaging dataset can be identified on the workstation. For finding structures not directly visible in the patients (like infiltrating malignant brain tumors or breast cancers during breast-conserving surgery or lumpectomy), this process can be reversed, and the tumor visible on the images can be found in patients as is. Navigation systems available to date require more time for registration to add to total surgery time. To decrease this time, the point-to-point registration can be replaced by laser registration or other fast video registration methods (Fig. 1.8) [35].
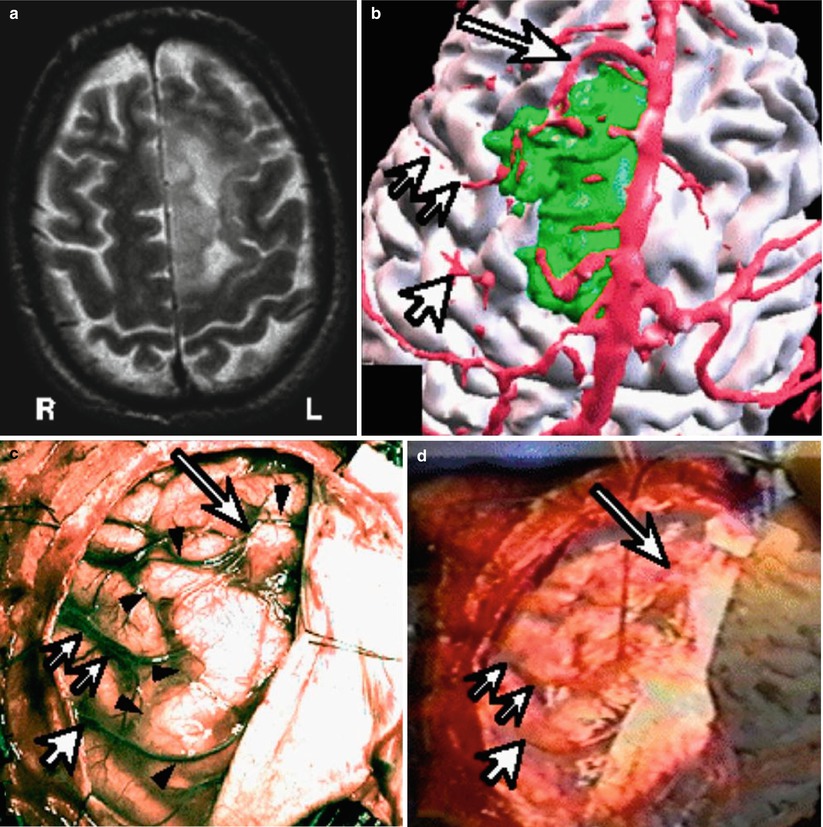

Fig. 1.8
Brain tumor resection with vessel to vessel video registration. (a) T2 weighted MRI of left sided brain tumor. (b) Segmented brain (white), vascular structures (red) and the tumor (green). (c) Intraoperative video of the brain surface, arrows showing one particular cortical vessel. (d) The segmented vessel overlayed on the videoimage of the real cortical vessel to improve registration (Nakajima et al. [34])
Image-guided surgery systems consist of hardware and software with the hardware including an image processing workstation interfaced with tracking technology. Tracking is the process by which interactive localization is possible in the patient’s coordinate system, and methods for tracking include articulated arms, active or passive optical tracking systems, sonic digitizers, and electromagnetic sensors. Active optical trackers use multiple video cameras to triangulate the 3D location of the flashing light-emitting diodes (LEDs) that may be mounted on any surgical instruments. Passive tracking systems use a video camera (or multiple video cameras) to localize markers that have been placed on surgical instruments. The infrared light reflected by these instruments is relayed to the workstation to calculate the precise location of the instrument in relation to the body organs in the surgical field. These systems use no power cable attached to the handheld localizer. Unfortunately, both LED and passive vision localization systems require line of sight between the landmarks or emitters and imaging sensors. Electromagnetic digitizers operate without using optical methods and can track instruments like flexible endoscopes, needles, guide wires, or catheters placed inside the body. Small electromagnetic transmitters are placed on instrument handles, allowing image-guided surgery systems to detect the position of the receiver holding the instrument in 3D space. The instruments are then displayed on a screen, even if a surgeon or instrument breaks the line of sight. Electromagnetic tracking alleviates some of the traditional line of sight problems encountered with optical localizers; however, it still has drawbacks. Large metal objects, however, distort the magnetic field and diminish accuracy. Nevertheless, electromagnetic navigation, which is virtually transparent to the surgeon, is the optimal tracking method used in IGT. Hybrid systems use both active and passive tracking. It is possible to combine optical and electromagnetic devices such as when the line of sight is broken. The electromagnetic tracking system can take over from the optical system.
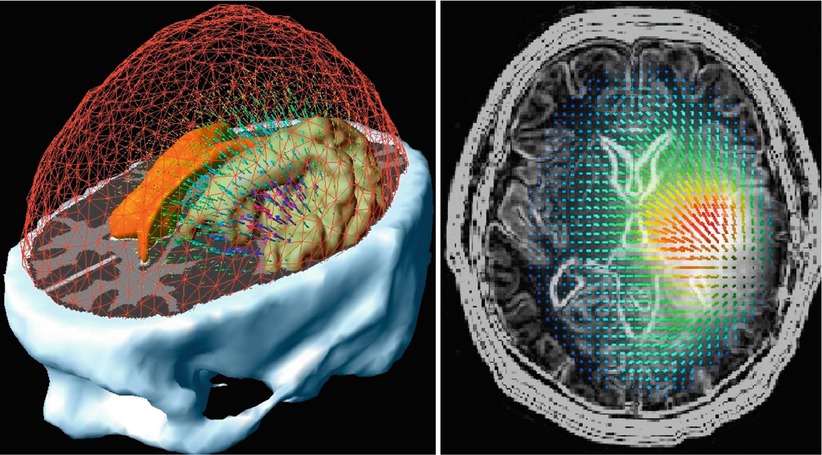
Advances in MRI permitted the further development of frame-based stereotactic neurosurgical procedures including biopsies and open surgeries. However, because of the inconvenience of frame-based systems, many neurosurgeons turned to frameless stereotactic systems. Frameless, computer-assisted navigational systems, however, require segmentation, registration, and interactive display. Navigation also requires appropriate displays that show the position, orientation, and trajectory of the surgical instruments within the patient’s dataset. Most displays incorporate both 2D and 3D data. Two-dimensional orthogonal displays generally show images from three different views: axial, coronal, and sagittal that are displayed simultaneously within the 3D display to allow the surgeon to navigate the instruments. For real-time navigation, interactive imaging is necessary, including the control of imaging planes interactively, a feat that is relatively easy to accomplish with handheld US transducers when the operator can freely change the plane of imaging. When MR image planes are changed interactively, this action requires more sophisticated control of image acquisition [36, 37]. Figure 1.9 shows the concept of interactive image acquisition as it was implemented in the first intraoperative MRI.
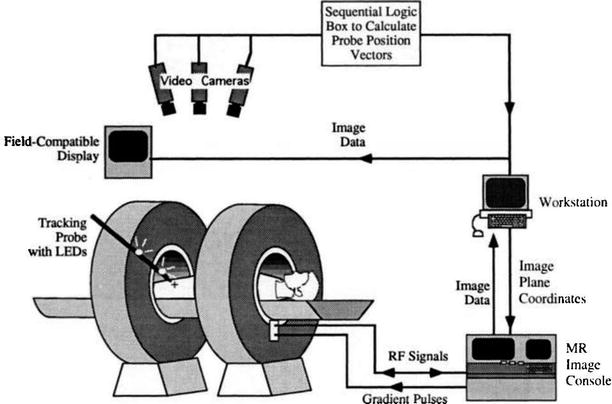

Fig. 1.9
Schema of interactive image control shows an open-magnet design that permits direct clinical access to the patient and simultaneous control of the MR imaging process (Schenck et al. [38])
Accuracy
Accuracy—the difference between the optimal and the realized correct solution—can be defined separately for registration, tracking, scanning, and clinical accuracy. Registration methods must be accurate and robust with both being important for assessing a given approachs. Robustness depends on the frequency of achieving the optimal solution. Accuracy and robustness are influenced by multiple factors and the lack of a gold standard. Human errors are the main factor affecting clinical accuracy. Technically, accuracy is influenced by image resolution, surgical equipment, and the type of surgical procedure. Accuracy is a fundamental prerequisite for all IGT systems that must provide clinical accuracy as good as, or better than, accuracy achieved without the system. The clinical accuracy of various image-guided surgical systems is converging into similar, acceptable ranges of a few mm that is acceptable in most applications.
Intraoperative Imaging
Surgical manipulations and maneuvers change the anatomical position of organs and soft tissues and lesions within them. For example, the leakage of cerebrospinal fluid (CSF) after opening the dura, hyperventilation, the administration of anesthetic and osmotic agents, and the retraction and resection of tissue all contribute to the shifting of the brain parenchyma [39]. Figure 1.10 shows displacement vectors due to mass effect and Figs. 1.11 and 1.12 show changes after craniotomy. If the patient is on the operating table in right lateral decubitus position, the organs shift from the position that they were in during the diagnostic exam done in the supine position. During laparoscopic surgery, large amount of CO2 gas is introduced, causing the displacement of abdominal organs that shift from their original position. These displacements make information from preoperatively acquired datasets unreliable at a substantially increasing rate as a procedure continues. Intraoperative imaging offsets this phenomenon by accurately updating images to account for these changes. The challenge of intraoperative imaging is not only to receive an update of the position of the deformed or displaced tissue but also to provide the interactivity required for an image-guided therapy system to follow and compensate for these changes [37]. Interactive intraoperative guidance allows one to accurately localize and target during surgery, to optimize surgical approaches that avoid critical structures, and to decrease vulnerability of surrounding normal tissues. Operating under interactive image guidance offers advantages over traditional guidance systems that use only preoperative data and, therefore, cannot provide accurate localization and navigation.
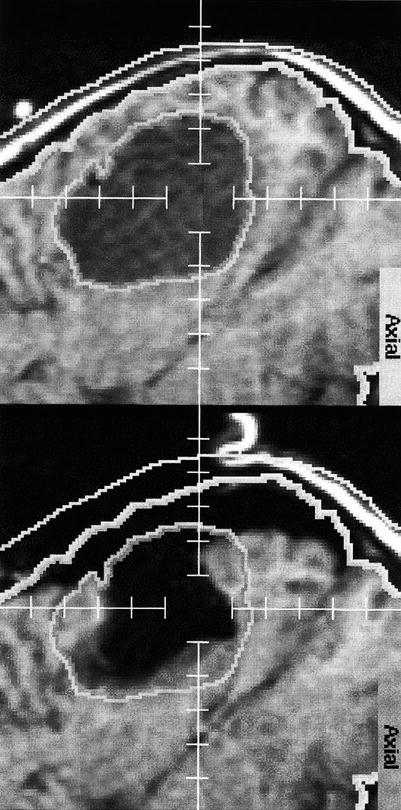
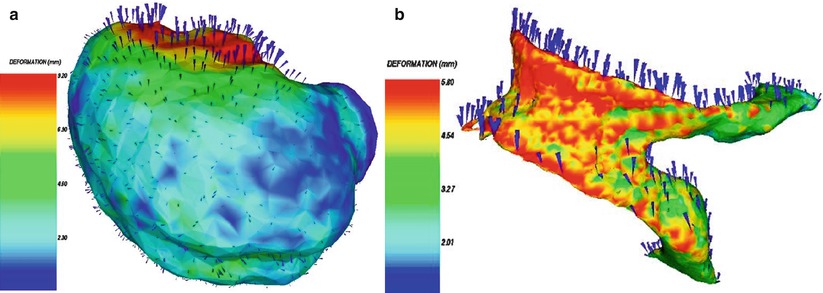

Fig. 1.11
Brain shift: The upper image shows the outlines/segmentation of an oligodendroglioma before craniotomy. The lower image shows the overlay of the same segmentation onto the corresponding post-resection image (Nabavi et al. [37])

Fig. 1.12
3D surface renderings of deformable surfaces with color-coded intensity of the deformation field. (a) Brain surface, (b) lateral ventricles (Ferrant et al. [23])
The other advantage of intraoperative imaging is the improved tissue characterization over direct surgical visualization. Most of the infiltrative malignant tumors are invisible to the surgeon, even though they are depicted by advanced intraoperative imaging modalities like MRI and PET/CT. These image-based information can be used to help the surgeon distinguish between normal from abnormal tissue during resection, and it offers superior capabilities in localizing a lesion in 3D spaces and allows this localization to be updated in a dynamic fashion, which has major implications in terms of the clinician’s ability to obtain accurate biopsies or correct resection margins. Interactive use of the intraoperative imaging modality allows precise maneuverings and the selection of the optimal trajectories for various surgical approaches. Brain tumor surgeries can be carefully planned and then executed under intraoperative MRI guidance [31, 41–43], to allow for surgical exposure and the related damage to the normal brain to be minimized. The maximal preservation of normal tissue may contribute to decreased surgical morbidity. The intraoperative MRI and other intraoperative imaging systems can help to decrease surgical complications through the identification of normal structures, such as blood vessels, white matter fiber tracts, and cortical regions with functional significance. Detection of possible intraoperative complications, such as hemorrhage, ischemia, or edema, is possible and can directly affect outcomes.
In the last two decades in addition to x-ray fluoroscopy and US, MRI has been introduced into the operating room [44, 45]. Cone-beam x-ray angio–fluoro systems are also installed in so-called hybrid operating rooms, in most cases, in combination with endovascular US. In a later phase intraoperative MRIs were combined with x-ray fluoroscopy (X-MR), and, more recently, a multimodality integrated operating room system, the AMIGO, was implemented at the Brigham and Women’s Hospital (BWH) that combines all imaging modalities, including PET/CT and optical imaging [46]. Intraoperative MRI dates back to the 1990s and, since then, has been successfully applied in neurosurgery for three primary reasons with the last one becoming the most significant today: (1) brain shift-corrected navigation, (2) monitoring/controlling thermal ablations, and (3) identifying residual tumor for resection [47]. There is a direct correlation between the extent of surgical resection and survival for neurosurgical patients who underwent surgery for low-grade glioma under intraoperative MRI guidance [48, 49]. Intraoperative MRI today is moving into other applications, including cardiovascular, abdominal, pelvic, and thoracic treatments. These procedures require advanced 3 T MRI platforms for faster and more flexible image acquisitions, higher image quality, and better spatial and temporal resolution; functional capabilities including fMRI and DTI; nonrigid registration algorithms to register pre- and intraoperative images; non-MR imaging improvements to continuously monitor brain shift to identify when a new 3D MRI dataset is needed intraoperatively; more integration of imaging and MRI-compatible navigational and robot-assisted systems; and greater computational capabilities to handle the processing of data. The BWH’s “AMIGO” suite is a setting for further progress in intraoperative MRI by incorporating other modalities including molecular imaging. Currently several IGT procedures are performed in the AMIGO such as neurosurgical procedures (open craniotomies for brain tumor resection, transnasal pituitary tumor removal, MRI-guided laser treatment of brain tumors); breast-conserving surgeries for breast cancer; ablation treatments of atrial and ventricular fibrillation; low-dose brachytherapy for prostate and high dose for cervical cancers; and MRI-, CT-, and PET-/CT-guided cryoablations of soft tissue tumors. Most recently image-guided parathyroid surgery and minimally invasive endoscopic removal of small lung cancers were introduced into AMIGO’s repertory.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree