Fig. 60.1
Published data on image-guided liver surgery and arranged by nature of end points (technical, clinical) vs. objectivity (qualitative and quantitative)
While demonstrating efficacy on a technical level can be achieved with rather small patient collectives, statistically valid improvements of clinical end points typically require large randomized and controlled trials. As can be seen in Fig. 60.1, to date no data is available that can objectively demonstrate clinical efficacy of image guidance technology when applied to open liver surgery. So far, available studies have demonstrated that technical accuracy is at a level where its application becomes serviceable to the surgeon [8–11]. Furthermore, surgeons who have been using those systems report information that is telling on a qualitative level, namely, that stress levels in the OR can be reduced due to improved planning and more precise in situ orientation [8, 9, 11]. Nevertheless, apart from a few individual cases that were reported as benefiting directly from the application of a CALS system [3] and one study that identified a slightly improved R1 resection rate when using instrument guidance [12], the clinical benefits of image guidance systems for open liver surgery need to be substantiated in the future with quantitative clinical data.
Instrument Guidance for Open Surgical Resection and Ablation
Navigated Resection
Image-based volumetry is clinically used to determine if a patient is eligible for surgical resection of the liver and to evaluate the risks involved in such a resection. Software for volume analysis forms part of most PACS systems available in modern hospitals. More advanced surgical planning software also helps to optimize resection proposals based on patient-specific information regarding liver blood supply (through hepatic arteries and portal veins) and drainage (through liver veins). The leading solution for such advanced planning is provided by MeVis Medical Solution (Bremen, GER) and provides a function analysis of different resection proposals (Fig. 60.2).
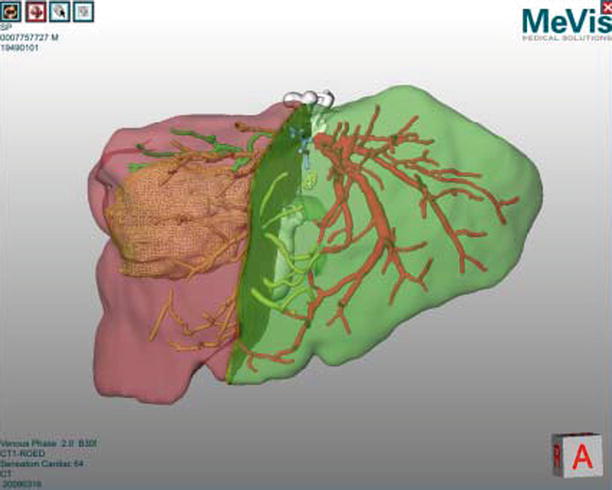

Fig. 60.2
Virtual liver surgery planning system (MeVis Medical Solutions AG)
Instrument guidance systems connect this planning stage with the actual surgical procedure and enable accurate reproduction of the planned resection planes in the OR. A combined visualization of the real-time tool positions in the coordinate system of the available image and planning information (Fig. 60.3) helps to improve targeting accuracy and situs orientation. Future improvements in resection guidance include dedicated system for aligning the resection instrument relative to the planned resection planes and modification of such plans based on intraoperative findings [13]. The main clinical indications for navigated resection are oncological resection cases as well as living donor transplantation procedures.
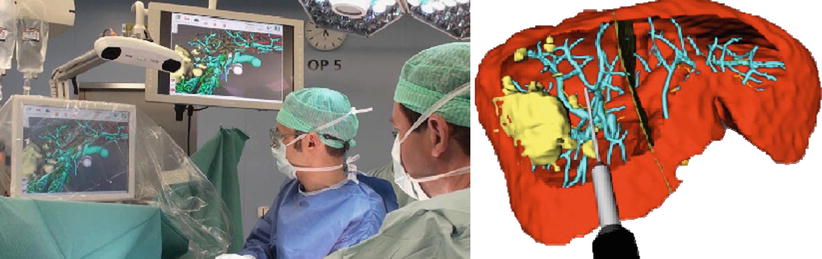

Fig. 60.3
Left: Intraoperative setup during image-guided liver surgery; Right: Screenshot of a navigation screen visualizing the tip of a resection instrument virtual resection plan (Peterhans et al. [11])
Navigated Ablation
Intraoperative ablation treatments require accurate placement of the applicator instrument for ensuring successful tumor ablation with sufficient safety margins. The use of soft tissue navigation technology has the potential to guide tools on a direct path towards tumors and to visualize risk structures that should not be damaged during needle insertion (Fig. 60.4 left). Typical functions during image-guided ablation are the progressive alignment of the ablation needle on a (planned) trajectory towards the tumor, visualization of the distance to the tumor center during the insertion, and previewing structures at risk along the insertion path. More advanced functionalities include a qualitative or quantitative prediction of the ablation volume, assisting the surgeon in achieving optimal safety margins, thus in predicting the effects of the treatment on vessels surrounding the tumor and on the liver capsule (Fig. 60.4 right). Basic ablation prediction uses the manufacturer’s specifications for a generic liver model while more sophisticated solutions use patient-specific simulations of heat propagation in the presence of cooling caused by vessels in the ablation zone [14].
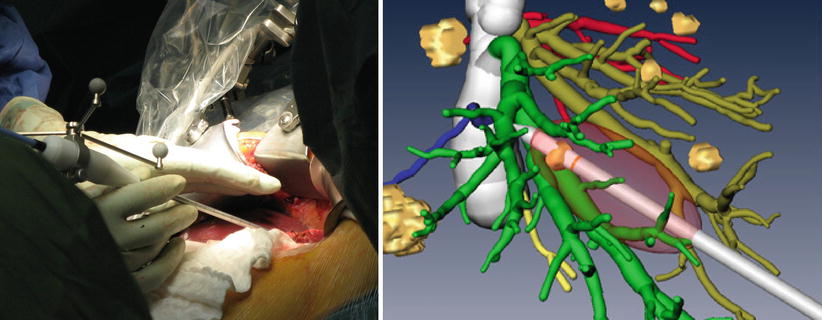

Fig. 60.4
Left: Navigated microwave ablation system. Right: Prediction of prospective ablation volume in the image-guided context of a 3D image and planning data set
Joint US and CT Visualization
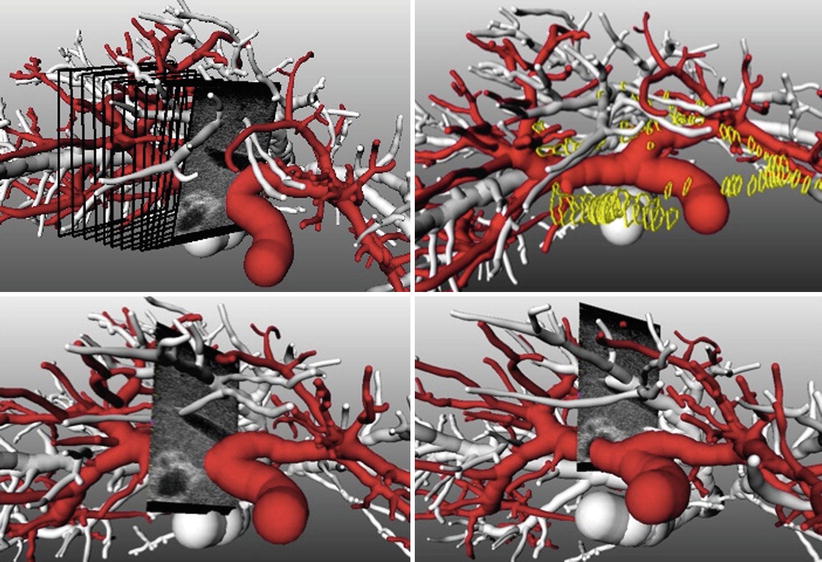
To gain improved understanding of the complex vascular anatomy during the actual surgery, surgical planning data can be visualized together with intraoperative ultrasound (hereafter US) imagery. While the ultrasound data is available in real time throughout the intervention, preoperatively acquired image and planning data needs to be co-registered to the patient and thus to the coordinate system of the ultrasound image data. Such a combined visualization and the effects of its application in a study of eight patients are described in [11]. If the joint visualization mode is enabled, the available 3D planning data (preoperatively) is rotated such that a perpendicular view of the US image is achieved. Automatic zooming allows for viewing the ultrasound at a constant resolution (Fig. 60.5 left). All extensive vascular structures are then cropped between the viewer and the ultrasound image (Fig. 60.5 right). Effectively, the surgeon benefits from an intuitive fusion of all available image modalities in the coordinate system of the navigation device.
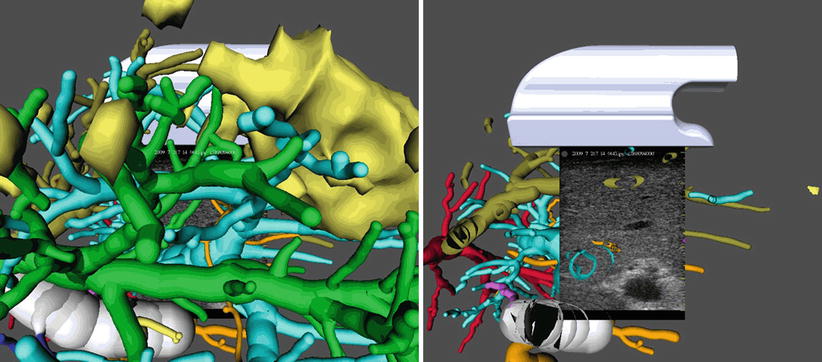

Fig. 60.5
Left: Virtual cameras adjustment for perpendicular view on the ultrasound imaging plane. Right: Complexity reduction by removing structures located between the virtual camera and the ultrasound plane
Risk Maps
The optimal interpretation of surgical planning data remains challenging, since a vast amount of data about the anatomy surrounding surgical activity needs to be taken into account. In order to reduce the complexity inherent in visually interpreting 3D contexts, a compact representation of surgically relevant information by means of a risk map has been proposed [15]. Such risk maps use color coding to highlight distances between vessels at risk and desired targets (such as tumors) and display them on the planned resection plane (Fig. 60.6).
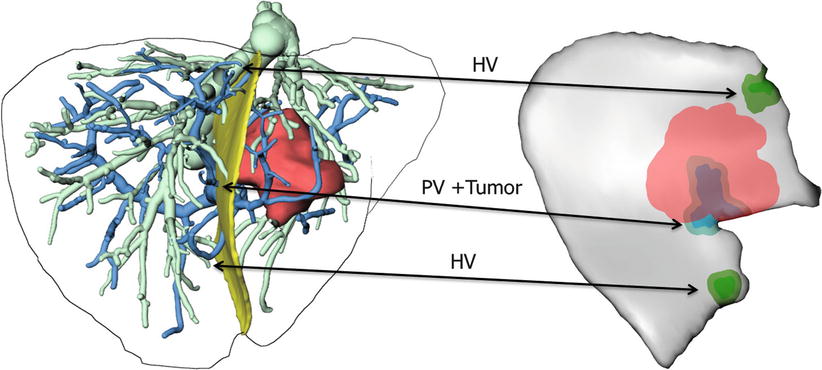

Fig. 60.6
Risk map for intraoperative application during instrument-guided procedures. Distances of 2 and 5 mm to hepatic veins (HV), portal veins (PV), and tumors are coded in different colors for each structure (Image courtesy: C. Hansen; MeVis Bremen, Germany)
Registration and Soft Tissue Tracking
In contrast to other surgical domains, such as orthopedic, neuro-, and ENT surgery, computer assistance in the domain of the soft tissues has to cope with an inherently unstable anatomical situation due to the elasticity and mobility of the target organ. Thus, the fundamental challenge in image guidance in open liver surgery is the registration of preoperatively acquired image data (3D/4D contrast-enhanced CT or MRI) with the intraoperative situation. This section discusses the different approaches that have been proposed by various groups and gives an overview of the current research towards registration methods that address the intrinsic challenges of open liver surgery.
Landmark-Based Registration
The use of corresponding landmarks between the patient and the image data sets has been transferred from other surgical domains to liver surgery. The major points that can be identified on the liver surface include vessel entrance points, the ligamentum falciforme and its fissure, prominent points on the edges of the liver, as well as tumors lying close to the organ surface. A first attempt in using such landmarks is reported in [16] and is later refined in a combined landmark-surface registration approach [17]. A clinical evaluation of the achievable accuracy and the intraoperative time requirements shows that landmark-based registration leads to a median target landmark registration error of 6.3 mm with a median time requirement of 1.5 min per registration update [11].
Surface-Based Registration
The acquisition of the entire organ surface leads to a significant increase of position data when compared to the single landmarks on the organ surface. However, the challenges of such approaches lie in efficiently acquiring the organ surface and in establishing correspondence. The use of laser surface scanning devices [1] leads to dense organ surface measurements but results in a large number of measurements from surrounding structures, which need to be removed manually. Due to non-unique point correspondences on a smooth, curved surface such as the liver, surface scans are used in combination with a set of corresponding landmarks [17].
Ultrasound-Based Registration
Intraoperative ultrasound imaging provides highly accurate real-time information on the position of internal structures of the liver. It is therefore expected that ultrasound-based registration will lead to a significant increase in registration accuracy compared to registration approaches based on surface data. The challenges in ultrasound-based registration lie in (a) the large variability of image quality during the acquisition and in (b) the extraction of relevant information when using such images for registration. Large variations in the resulting data quality are caused by imaging settings, correlation between sound propagation parameters and tissue density, and user-dependent imaging protocols in the case of free-hand ultrasound. This last effect can be minimized by guiding the user during the scanning process as illustrated in Fig. 60.7 or through the use of 3D imaging equipment [12].
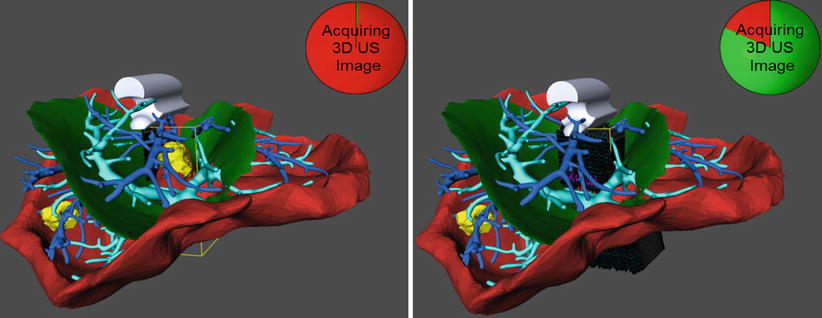

Fig. 60.7
Interactive recording of an ultrasound sweep in an initially rigidly registered situation. Left: The user places the probe as desired and triggers the recording of a set of images within a defined geometric frame (yellow bounding box) and within a certain amount of time. Right: The circle clock visualizes the state of completion of the recording. A 3D ultrasound volume is then reconstructed and used to refine the initial registration
Once an intraoperative ultrasound data set is acquired and found to be of good quality, correspondence between ultrasound measurements and structures in the preoperative image data sets needs to be established. The most prominent structures that can be detected in both image data sets are the different liver vessel systems. The following three paragraphs describe approaches for using vessels detected in ultrasound images to improve the registration process.
Landmark-Based: Vessel bifurcations are used for orientation references in conventional liver surgery and can be localized with a high degree of accuracy in both ultrasound and CT/MRI data sets. Automatic detection of such points is explored in [18] but has not led to a clinical application. The use of manually selected vessel landmarks is illustrated in Fig. 60.8. A systematic evaluation of such an approach, i.e., with rigid and nonrigid registration, shows a mean accuracy of 2.6–4.9 mm with a time overhead of 5–10 min per registration update [19].
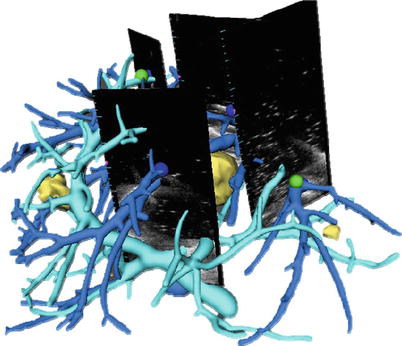

Fig. 60.8
Rigid registration using vessel-based landmarks
Contour-Based: When compared to landmark-based approaches, the automatic extraction of vessel contours from ultrasound images [20] provides additional information about the diameter. Based on this parametric vessel representation, a registration algorithm using vessel diameter and centerline information increases the registration robustness by checking possible corresponding landmarks for distance and vessel diameter constrains [21]. Initial results (Fig. 60.9) show an improvement in registration accuracy when compared to landmark-based registration using surface points.