(1)
School of Medicine and Biology, St. Andrew University, Scotland, UK
Abstract
Immunoelectron microscopy (immuno-EM) using gold labeling on sections is a powerful technique for mapping the distribution of proteins, lipids, carbohydrates, and nucleic acids in intact biological systems. The gold particles comprise a useful and readily quantifiable digital readout. Simply applying a labeling reagent (antibody or other affinity probe) to an ultrathin section yields a pattern of gold signal over the biological structures displayed in the section. This initial (raw) distribution of gold signal contains both specific and nonspecific labeling. Here we describe a method for removing nonspecific labeling to leave the target-specific signal. This specific labeling distribution better reflects the “real” distribution of the cell component of interest.
Key words
StereologyKnockdownKnockoutImmunoelectron microscopyQuantitationQuantificationSpecificityGold labelingImmunogoldSpecific labeling1 Introduction
Immunoelectron microscopy using colloidal gold particles maps target components in the complex array of cell compartments and structures displayed in ultrathin sections [1]. Sectioning opens up compartments and presents them to the labeling system, while the gold signal represents a digital readout that is easily quantifiable. Over the past few decades, an array of methods have been developed for quantifying gold particles on sections [2–4]. These methods can be combined with appropriate sampling to allow unbiased and efficient readouts of gold particle concentration (density) over cellular structures or of the gold particle distribution over a series of compartments/structures.
Gold labeling in immuno-EM is composed of two key quantities: (a) specific labeling of target components to which the labeling is directed in the first place and (b) nonspecific labeling which represents interactions of the labeling system with nontarget components and brings noise to the system [5]. Despite the widespread use of immunogold EM, the labeling is often quantified without further analysis of the specificity. However, there is now a wide range of established methods for modifying the expression of proteins in situ, and these provide opportunities for estimating the fraction of the labeling that represents the component of interest (e.g., see [6–8]).
The principle described here is best explained using the knockdown/knockout approach in which two samples are prepared [5]: one to which no treatment has been applied and the other in which there has been a reduction in the expression of the target component. The two samples are then fixed, embedded, and sectioned before labeling with the affinity probe of interest and the colloidal gold reagent. Gold labeling is carried out under standardized conditions and as far as possible using the same solutions.
Using established methods for quantification, the first step is to estimate labeling density over compartments of interest under the two conditions. The densities may be expressed as gold particles per unit profile area or membrane profile length, each of which can be estimated stereologically. The first key indicator of specificity is the difference in the density of labeling between the untreated and the treated (knockdown/knockout) sample. This density drop provides an estimate of the specific labeling density. The next step is to calculate the fraction of labeling that is specific by comparing the change in labeling density with the gold signal obtained under conditions of initial expression. If the fraction specific is, for example, 0.8, then each gold particle has an 80 % chance that it labels the target component under the initial condition. Finally, it is possible to correct the initial labeling distribution by removing nonspecific background signal. This is done by multiplying the fraction specific and the number of gold particles counted for each compartment in the initial condition. In the case of complete knockdown, the specific labeling distribution will reflect the distribution of available molecules in the sample. If a knockdown is partial, then the specific labeling will represent that fraction of the component.
This same principle can be applied to labeling reagent controls such as peptide inhibition (see example below), a, albeit, less rigorous way of controlling specificity. An additional approach to establishing specificity is when expression in the sample has been enhanced (by expressing tagged protein, novel targets, or uptake of exogenous molecules). Here the strategy is to calculate the fraction specific by comparing the change in labeling density with the signal density after enhancement (again see below).
2 Materials
1.
Gold-labeled ultrathin sections from untreated specimen and the specimen with reduced/enhanced expression.
2.
Standard transmission electron microscope with image capture capability such as a digital camera.
3.
Stereology or imaging-processing software such as Adobe Photo-shop for generating a systematic grid lattice.
3 Methods
3.1 Processing, Sectioning, and Labeling (Guidelines Only)
1.
Process the two samples for immunoelectron microscopy. In the case of reduced expression, one specimen is processed for the gene knockout or knockdown of expression (see Note 1 below). In the case of enhanced expression, the component might be introduced by microinjection or by a physiological process such as endocytosis or by expression of untagged or epitope-tagged target. At this stage it is helpful to ensure as little variation in the target component expression/knockout/deletion as possible. This will allow overall estimates for the experiments/animals/cultures to be computed. For controls involving peptide inhibition, it is important to take sections for the two conditions from the same sample blocks.
2.
Fix the organisms/cells preferably with glutaraldehyde-containing fixatives. Then embed in resin or freeze in cryoprotectant (thawed frozen sections) according to the optimal method for preserving the structures and providing a good yield of immuno-label. Pilot experiments are needed to obtain this information and to determine the best fixation conditions. In general thawed frozen sections provide the best morphological display of membranes and most intense immunogold signals. High-pressure frozen material embedded in resin is also suitable.
3.
Cut and mount ultrathin sections on support grids. For specimen-based studies, sections from both control and enhanced/reduced expression samples should be cut and processed in parallel. When reagent-based controls are used (e.g., peptide inhibition), the sections should be taken from the same block and processed identically as far as possible.
4.
Label sections using predetermined concentrations of affinity reagent. The conditions should be selected as those that provide a maximum difference in the labeling intensity between the full expression and the reduced expression condition.
3.2 Quantification and Computation of Specificity Indices
1.
Choose a magnification at which both the gold and the compartment structure profile of interest can be identified easily. For this purpose a gold size of 8 nm is recommended. This choice is a trade-off between a larger gold size (that allows lower magnification to be selected) and a smaller gold size (that confers an enhanced labeling signal). It is usually more efficient to reduce the magnification to include as much relevant structure as possible.
2.
Select a section with minimal number of folds in which the structures are clearly displayed. Record images by systematic uniform random (SUR) sampling [9]. To do this, an electron microscope grid-square corner outside the section is a convenient random starting point, and from this location, micrographs are taken in a systematic array covering the whole area of the section. A useful starting point is 20 micrographs. Micrographs are only recorded from the array if they contain a feature of interest.
3.
Get Clinical Tree app for offline access

Estimate the area or the profile membrane length of the compartments of interest within the micrographs as appropriate. Area is used when the gold labeling is located over the volume of an organelle exposed by the section, and the length of membrane profiles is used when the gold is located at or over membrane profiles is used when the target is located at or within the membranes. A mixture of gold densities over area and length for different organelles is allowed when assessing specificity (see [4]). Area estimation is carried out using point counting with a randomly placed systematic lattice of points. Use a lattice density, which will give a total of approximately 100–200 point hits per section over all organelle profiles in the first instance. The sum of point hits over the organelle of interest (ΣP) multiplied by the area associated with a point (a) is an estimate of the aggregate area (A):
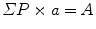
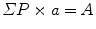
In Adobe Photoshop, square grid lattices can be generated using the grid function in this software. The total number of point hits can be regulated by varying the spacing between the points. It is usually prudent to draw a quadrat surrounded by a guard area. This allows clear identification of structures near the quadrat edge especially when their profiles extend into the guard area. Counts of gold particles associated within each compartment located within the quadrat are made (ΣΝg). The labeling density is calculated from
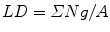
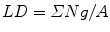
If profile length (B) is required, the appropriate formula would be
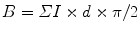 where ΣI is the sum of intersections of test lines with the membrane profiles of interest, d is the spacing between the lines, and π/2 is the constant applied when parallel test lines are used (if lines of a square lattice grid are used, then the constant would be π/4). Again in the first instance, the aim is to obtain a total of approximately 100–200 intersection hits per section over all organelle profiles (see Notes 2 and 3 below).
where ΣI is the sum of intersections of test lines with the membrane profiles of interest, d is the spacing between the lines, and π/2 is the constant applied when parallel test lines are used (if lines of a square lattice grid are used, then the constant would be π/4). Again in the first instance, the aim is to obtain a total of approximately 100–200 intersection hits per section over all organelle profiles (see Notes 2 and 3 below).
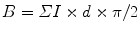
(a)
Specific labeling density, LD(sp), is then given by calculating the difference between the initial (raw) labeling density, LD(0), and the labeling density after reduced expression, LD(−) as follows:
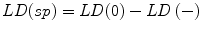
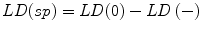
(b)




Fraction specific, F(sp), is given by
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree