Fig. 14.1
Main features of the different imaging biomarkers platform architectures existing nowadays
14.4 A Proposed Architecture
The ideal solution would consist on a platform covering the features mentioned in the previous section and being able to be installed not only in the cloud but also in local facilities at hospitals. Furthermore, it should be oriented not only to medical doctors as users but also to any researcher in medical imaging quantification and also to clinical research organizations (CROs) managing clinical trials.
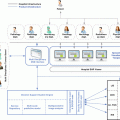
For these purposes, a modular architecture must be one of the most important requirements in order to take advantage of as much modules as possible for either cloud or local solutions (see Fig. 14.2).


Fig. 14.2
Imaging biomarkers analysis local and cloud platform sharing the algorithms for image analysis for both architectures
For this structure, a database was engineered taking into account all entities and relationships present in the imaging biomarkers platform. The database considers the imaging studies, the processes of imaging biomarkers analysis launched, their status (not started, in process, error, finished), the results, the institution and equipment of the users, and the reference values for healthy populations. The database structure can be appreciated in Fig. 14.3.


Fig. 14.3
Design of the results hierarchy for the imaging biomarkers analysis platform
Regarding the local solution to work within the radiological workflow, a detailed diagram of the local solution can be appreciated in Fig. 14.4.


Fig. 14.4
Detailed architecture of the local platform for imaging biomarkers analysis
A DICOM node is used to continuously “listen” for any incoming data. It can be configured in the systems of the network by its IP number, AE title, and port number. The node allows for the reception of imaging studies either from the PACS or directly from the equipment. If appropriate rules are set by the PACS provider, it is possible to auto-push studies from the PACS to the imaging biomarkers’ DICOM node as they are received, specially for those analysis pipelines that are fully automatic (i.e., brain volumetry, multiple sclerosis lesion segmentation, among others). The node performs two main tasks: stores the imaging data in a standardized way in the local server for imaging biomarkers analysis and introduces a new registry in the platform database. The DICOM node is also responsible for the notifications to the users managing the platform.
The web application is the core of the solution; all the logics in the application is checking for the entry of new studies by queries to the database and deleting the source images of those studies stored for more than 1 month. The application is responsible for launching processes of imaging biomarkers analysis. For that, the imaging biomarker solutions have been implemented by following an internally standardized format, which consists in specifying the route with the source images, the route where the results must be stored, the process ID, and some biomarker configuration settings that the user has selected in the application before pushing the button for the analysis. The application is also checking for the existence of errors in any of the running processes either in Python, MATLAB, or in the system. The interfaces of the web application are built based on the HTML5, CSS3 and JavaScript standards, allowing a rich and friendly usability. For this propose, and considering the characteristics of the product and the user needs, AngularJS has been selected as the most suitable framework. These interfaces create a client-side environment that allow intuitive interactions and fast communications with a server-side build using Java. The communication between server (backEnd) and client (frontEnd) is made via API-REST (Representational State Transfer), so a SPA (Single Page Application) can be built allowing to show many different views with a small amount of data browsing through the web via structured JSON objects. This methodology implies that other Web applications are able to use API-REST to connect with the database.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree