Fig. 1
The feature space is defined by the cross-section center position x = (x 1, x 2, x 3), the cross-section tangential direction Θ = (θ 1, θ 2, θ 3) and the lumen pixel intensity distribution p vessel
2 Segmentation Model & Theoretical Foundations
2.1 Vessel Model & Particle Filters
To explain our method at a concept level, let us assume that a segment of the vessel has been detected: a 2D shape on a 3D plane. Similar to region growing and front propagation techniques, our method aims to segment the vessel in adjacent planes. To this end, one can consider the hypotheses ω of the vessel being at a certain location (x), having certain orientation (Θ), and referring to certain shape – an elliptic model is a common choice (є) – with certain appearance characteristics (p vessel ).
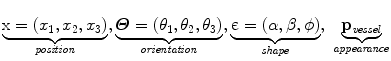
Then, segmentation consists in finding the optimal parameters of ω given the observed 3D volume. Let us consider a probabilistic interpretation of the problem with π(ω) being the posterior distribution that measures the fitness of the vector ω with the observation. Under the assumption that such a law is present, segmentation consists in finding at each step the set of parameters ω that maximizes π(ω). However, since such a model is unknown, one can assume an autoregressive mechanism that, given prior knowledge, predicts the actual position of the vessel and a sequential estimate of its corresponding states. To this end, we define:
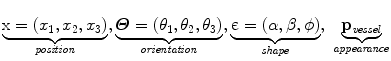
(1)
an iterative process to predict the next state and update the density function, that can be done using a Bayes sequential estimator and is based on the computation of the present state ω t pdf of a system, based on observations from time 1 to time t z 1:t : π(ω t |z 1:t ). Assuming that one has access to the prior pdf π(ω t−1 |z 1:t−1), the posterior pdf π(ω t |z 1:t ) is computed according to the Bayes rule:
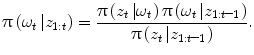
a distance between prediction and actual observation, based on the observation.
Simple parametric models will be suceptible to fail with vessels’ irregularities (pathologies, prosthesis, …). Therefore instead of optimizing a single state vector, multiple hypotheses are generated and weighted according to actual observation. Nevertheless, in practical cases, it is impossible to compute exactly the posterior pdf π(ω t |z 1:t ). An elegant approach to implement such a technique refers to the use of particle filters where each given hypothesis is a state in the feature space (or particle), and the collection of hypothesis is a sampling of the feature space.
Particle Filters [1, 8] are sequential Monte-Carlo techniques that are used to estimate the Bayesian posterior probability density functions (pdf ) [16, 34]. In terms of a mathematical formulation, such a method approximates the posterior pdf by M random measures {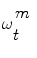 , m = 1..M } associated to M weights {
, m = 1..M } associated to M weights {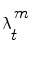 , m = 1..M }, such that
, m = 1..M }, such that
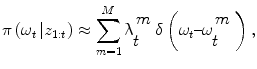
where each weight 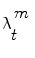 reflects the importance of the sample
reflects the importance of the sample 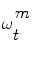 in the pdf. The samples
in the pdf. The samples 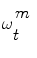 are drawn using the principle of Importance Density [9], of pdf
are drawn using the principle of Importance Density [9], of pdf 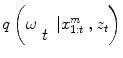 and it is shown that their weights
and it is shown that their weights 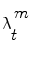 are updated according to
are updated according to
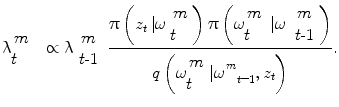
Once a set of samples has been drawn, 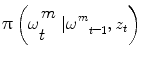 can be computed out of the observation z t for each sample, and the estimation of the posteriori pdf can be sequentially updated.
can be computed out of the observation z t for each sample, and the estimation of the posteriori pdf can be sequentially updated.
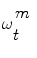 , m = 1..M } associated to M weights {
, m = 1..M } associated to M weights {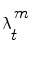 , m = 1..M }, such that
, m = 1..M }, such that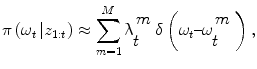
(2)
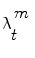 reflects the importance of the sample
reflects the importance of the sample 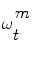 in the pdf. The samples
in the pdf. The samples 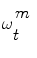 are drawn using the principle of Importance Density [9], of pdf
are drawn using the principle of Importance Density [9], of pdf 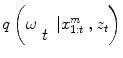 and it is shown that their weights
and it is shown that their weights 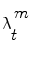 are updated according to
are updated according to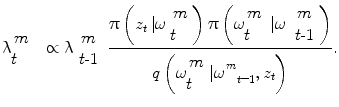
(3)
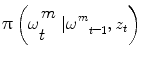 can be computed out of the observation z t for each sample, and the estimation of the posteriori pdf can be sequentially updated.
can be computed out of the observation z t for each sample, and the estimation of the posteriori pdf can be sequentially updated.2.2 Prediction & Observation: Distance
This theory is now applied to vessel tracking. Each one of the particles 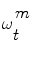 represents a hypothetic state of the vessel; a probability measure
represents a hypothetic state of the vessel; a probability measure 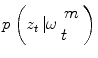 is used to quantify how the image data z t fits the vessel model
is used to quantify how the image data z t fits the vessel model 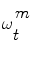 . To this end, we are using the image terms, and in particular the intensities that do correspond to the vessel in the current cross-section. The vessel’s cross-section is defined by the hypothetic state vector (see Eq. (1)) with a 3D location, a 3D orientation, a lumen’s diameter and a pixel intensity distribution model (the multi-Gaussian). The observed distribution of this set is approximated using a Gaussian mixture model according to the Expectancy-Maximization principle. Each hypothesis is composed by the features given in Eq. (1), therefore, the probability measure is essentially the likelihood of the observation z, given the appearance A model. The following measures (loosely called probabilities) are normalized so that their sum over all particles is equal to one. Assuming statistical independence between shape S and appearance model A, p(z t |ω t ) = p(z t |S)p(z t |A).
. To this end, we are using the image terms, and in particular the intensities that do correspond to the vessel in the current cross-section. The vessel’s cross-section is defined by the hypothetic state vector (see Eq. (1)) with a 3D location, a 3D orientation, a lumen’s diameter and a pixel intensity distribution model (the multi-Gaussian). The observed distribution of this set is approximated using a Gaussian mixture model according to the Expectancy-Maximization principle. Each hypothesis is composed by the features given in Eq. (1), therefore, the probability measure is essentially the likelihood of the observation z, given the appearance A model. The following measures (loosely called probabilities) are normalized so that their sum over all particles is equal to one. Assuming statistical independence between shape S and appearance model A, p(z t |ω t ) = p(z t |S)p(z t |A).
Get Clinical Tree app for offline access
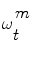 represents a hypothetic state of the vessel; a probability measure
represents a hypothetic state of the vessel; a probability measure 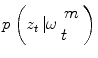 is used to quantify how the image data z t fits the vessel model
is used to quantify how the image data z t fits the vessel model 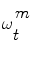 . To this end, we are using the image terms, and in particular the intensities that do correspond to the vessel in the current cross-section. The vessel’s cross-section is defined by the hypothetic state vector (see Eq. (1)) with a 3D location, a 3D orientation, a lumen’s diameter and a pixel intensity distribution model (the multi-Gaussian). The observed distribution of this set is approximated using a Gaussian mixture model according to the Expectancy-Maximization principle. Each hypothesis is composed by the features given in Eq. (1), therefore, the probability measure is essentially the likelihood of the observation z, given the appearance A model. The following measures (loosely called probabilities) are normalized so that their sum over all particles is equal to one. Assuming statistical independence between shape S and appearance model A, p(z t |ω t ) = p(z t |S)p(z t |A).
. To this end, we are using the image terms, and in particular the intensities that do correspond to the vessel in the current cross-section. The vessel’s cross-section is defined by the hypothetic state vector (see Eq. (1)) with a 3D location, a 3D orientation, a lumen’s diameter and a pixel intensity distribution model (the multi-Gaussian). The observed distribution of this set is approximated using a Gaussian mixture model according to the Expectancy-Maximization principle. Each hypothesis is composed by the features given in Eq. (1), therefore, the probability measure is essentially the likelihood of the observation z, given the appearance A model. The following measures (loosely called probabilities) are normalized so that their sum over all particles is equal to one. Assuming statistical independence between shape S and appearance model A, p(z t |ω t ) = p(z t |S)p(z t |A).
Probability measure for shape based on contrast
Given the vessel model (see Eq. (1)), whose parameters are specified by the particle ω t , a measure of contrast, that we call the ribbon measure R, is computed:
The probability of the observation given the shape model is then computed:
![$$ \left\{\begin{array}{ll} R=-\infty,&{\mu}_{\mathit{int}}\le {\mu}_{\mathit{ext}} \\[4pt] R=\frac{\mu_{\mathit{int}}\mathit{\hbox{--}}{\mu}_{\mathit{ext}}}{\mu_{\mathit{int}}+{\mu}_{\mathit{ext}}},& \mathit{otherwise} \end{array}\right. $$](/wp-content/uploads/2016/09/A151032_1_En_14_Chapter_Equ4.gif)
(4)
where R 0 is a normalizing constant (the average value of R from ground truth), μ int is the mean intensity value for the voxels in the vessel, and μ ext is the intensities mean value for the voxels in a band outside the vessel, such that the band and the vessel’s lumen have the same area. This measure is normalized to be equivalent to model a probability measure. Since the coronary arteries are brighter than the background, the best match maximizes R.
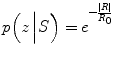
(5)
Probability measure for appearance
For the vessel lumen pixels distribution p vessel Eq. (1), the probability is measured as the distance between the hypothesized distribution and the distribution actually observed.
The distance we use is the symmetrized Kullback-Leibler distance D(p, q) between the model p(x) = p vessel and the observation q(x):