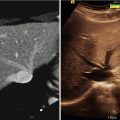
Fig. 4.1
The skull is shown by simple thresholding of CT volume (left); prostate segmentation from TRUS vibro-elastography volume (center); and a mandible segmentation from a curved CT cross section (right)
In certain image modality and anatomy combinations, segmentation can be a very difficult task, involving knowledge about the anatomy, the particular patient, the disease, the imaging modality, and sometimes even the imaging device. In most of the cases, the acquired image data are not sufficient to solve the segmentation problem, making it necessary to incorporate some form of prior information as well. Different techniques have been developed to represent such priors, many of them extending on methods derived from probability theory, where an effective segmentation strategy can be learned, e.g., from several observed segmentations. Statistical shape models [6] can further provide probabilistic estimates of anatomical structures and their variability based on large sets of previous segmentations, therefore improving segmentation outcome based on this prior information. Some other common methods for segmentation involve neural networks, which simulate the human learning process, and the use of graph theory, where pixel connectivity is represented as a graph. Figure 4.1 (right) shows an intensity-based automatic segmentation of a (curved) mandible cross section. Note that such probabilistic and statistical methods require an essential phase of model training, which may influence the segmentation outcome to a great extent.
Registration can be defined as the task of finding a mapping that aligns anatomical structures between two images [1, 2]. The nature of misalignment to be corrected for can be rigid, e.g., in case of bones since they do not easily deform, or deformable, e.g., brain-shift during surgery where different locations in the brain may displace by different amounts. Depending on the expected nature of misalignment in a particular task and also the affordable computational and implementation complexity, a mapping is then sought accordingly at a chosen level of sophistication, ranging from simple, pure translations or affine transformations to deformation fields, in which displacement is defined individually at each image location [7–9].
The nature and the cause of the misalignment can be diverse. These include temporal differences between image acquisitions, such as organ motion due to breathing or the state of bladder fullness; spatial differences in the positioning of the imaging device, such as in compounding for 3D ultrasound reconstruction from multiple 2D images acquired while moving the ultrasound transducer; the need to fuse images acquired by different imaging devices, such as MRI to PET registration; and sometimes differences in anatomical detail, which is unavoidable during matching images from different patients (inter-patient registration) or may likely happen due to significant changes in an individual patient’s anatomy, e.g., due to tumor growth or shrinkage. In most registration problems, however, a combination of these abovementioned phenomena does coexist.
Based on the inherent representation of the anatomy, registration methods can be classified into feature–based methods involving aligning manually or automatically selected anatomical points of interest; intensity–based methods determining a mapping by comparing image intensities or information derived thereof; and surface–based methods employing implicit/explicit models of anatomical structures for alignment. Of course, different combinations of the above are also possible, such as registering surfaces to boundary features. The success of an alignment is quantified by an appropriately defined similarity measure. This can for instance be the correlation of overlapping pixel intensities or the distance between registered surfaces, depending on the considered anatomical representation given above. Subsequently, a large class of registration methods involves the definition of such a similarity measure (aggregated over the entire image) as their objective function and making use of well-established optimization techniques for finding the mapping that maximizes the similarity between the images according to that chosen objective function. Several flavors of other techniques exist for registration and are further discussed in detail later in Chap. 3. Figure 4.2, on the left, shows the displacement of the liver during breathing obtained by the registration of MR images during the breathing cycle [10]. On the right of this figure, an inter-patient b-splines-based registration of corpora callosa from 2D MR images can be seen.
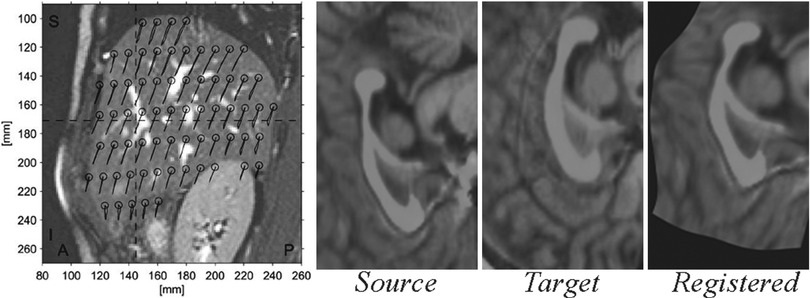

Fig. 4.2
Liver motion during breathing cycle derived by image registration (left) (Reprinted from von Siebenthal et al. [10]) and a diffeomorphic registration of corpora callosa using b-splines (right)
For both segmentation and registration, iterative methods exist; but these commonly require initialization, depending on which the results may significantly deteriorate when it is not sufficiently close to the sought optimum. In contrast, global search strategies, e.g., graph-cut methods [11], do not necessitate an initial state but often at the expense of large storage space or computational cost. In addition, for certain tasks that involve the application of both segmentation and registration consecutively, methods that treat them as a single, simultaneous task have also been proposed and successfully employed in the literature [12].
Localization Tools
Localization can be defined as discovering or determining the relative or absolute positions of certain targets in a surgical scene. Such targets can belong to the patient anatomy, as well as be parts of medical tools. A common method is to use natural anatomical landmarks in this task; nonetheless, artificial (fiducial) markers are also frequently utilized in the absence of suitable natural landmarks or in order to complement them. The accuracy of localization may affect, and therefore is essential for, a multitude of other succeeding computational processes, such as registration, referencing, scene augmentation, or navigation.
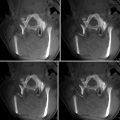
When localization is performed using medical imaging modalities, it relates closely to segmentation, since finding landmarks/markers will likely involve segmenting them from their background. Among other common techniques are optical and magnetic trackers, which require specialized devices and markers for accurate and/or convenient localization. Optical trackers use specific geometric patterns (like checkerboards) or infrared sources as markers and employ triangulation to compute 3D positions. They can offer high accuracy but may suffer from occlusion, limiting the operator’s movement. Magnetic trackers can detect the position and also orientation of their remote sensors with respect to a base device from the induced current on orthogonally placed coils inside them. While, on the one hand, they may suffer from limited field of view, possible interference with ferromagnetic surgical tools/devices, and typical lower accuracy compared to optical trackers; on the other hand, they do not require a visual line of sight and thus are commonly used in various diagnostic and therapeutic situations. A comparison of these two tracking systems was studied in a neurosurgical setup in [13]. The left image in Fig. 4.3 shows a magnetic tracker attached to an ultrasound probe, using which the spatial locations of the individual acquired frames can be inferred, allowing them to be placed in a common 3D reference frame as shown in the right image.
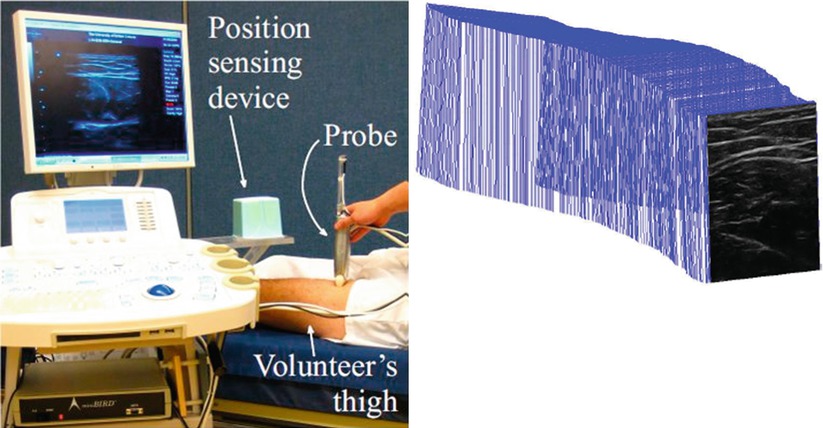

Fig. 4.3
An ultrasound transducer tracked by a magnetic position sensor (left); and ultrasound images that are acquired sequentially but placed in a common 3D reference frame based on the tracking data (right) (© [2011] IEEE. Reprinted, with permission, from Goksel and Salcudean [14])
Computer Simulation
Simulation is a procedure for replicating reality to some degree in a suitable (real or virtual) environment. The level of realism that a simulation needs to provide depends on the simulation objectives, the task and the environment being simulated, and the affordable simulation complexity. For instance, mannequins can be of great use in learning the basics of cardiopulmonary resuscitation, but may not be of as much help in training complex neurosurgical procedures. With the breathtaking development of available computational power, rather complex environments and tasks can now be replicated as virtual models, allowing the simulation of their behavior on a computer. Such simulations are achieved using a range of different methods, from simple to sophisticated; while some may have certain simulation aspects more emphasized and more realistic in comparison to others.
A computer simulation requires a model of the reality that is being simulated, and will output a state or response of that model based on the inputs provided, such as imagery or sensory data, or user interaction. Such models approximate reality in mathematical and algorithmic terms and can cover different aspects of their real counterparts, as in:
Their visual appearances
Their geometrical and/or mechanical properties
Their physiological behavior by accounting for the underlying biophysical, biochemical, and biological processes
Computer simulations are commonly used in planning interventions as well as assessing possible outcomes. Planning can be merely in the form of physical access to certain surgical locations and thus involve geometrical considerations alone. Nonetheless, in many occasions planning refers to the assessment of procedure outcome (efficacy) given multiple surgical variables and patient-specific anatomical and physiological conditions. Accordingly, planning necessitates a substantial amount of computational support and often formal optimization procedures for decision making.
An important aspect of simulation procedures is the computational time necessary to perform them. In some cases, as in surgical training simulations, real-time interaction is required necessitating rapid simulation cycles. In most of the image-processing scenarios, however, simulations can be performed offline, without the need for immediate feedback to the clinicians.
The output of a simulation can be numerical such as the dose at a location during radiotherapy. It can also be a visual rendering of the relevant anatomical structures, let’s say after an organ deformation to be caused by a tool interaction that the surgeon is anticipating to perform. In haptic training simulations for medical procedures, one of the (main) outputs is the simulated kinaesthetic force feedback that is estimated to act on the surgical tool due to contact with the virtual patient and, accordingly, applied on the user’s hand via a haptic device. Often accompanied by a virtual reality rendering, this force feedback is then applied on the user’s hand for a fully realistic immersion of the trainee in the simulation realm. During the past decade, the availability of supercomputing resources and the fast increase of our quantitative knowledge about physiology have led to the creation of increasingly complex models of healthy and pathological behavior of nearly all organs of the human body. Several large initiatives are currently working toward this goal, such as the Physiome [15] or the Virtual Physiological Human [16] projects.
Model Generation
Computational procedures rely on models of anatomical objects and physiological processes under investigation as an abstraction and semantic interpretation, in order to measure, simulate, and visualize them in the relevant medical context. A model may involve not only the appearance or the geometry of an organ but also its physiological properties, either (1) in an explicit form like the kinematics of motion of a joint or the movement and deformation of an organ due to respiration or (2) implicitly as a collection of parameters characterizing the involved tissues and their behavior, for example, the elastic properties of deformable organs to model their reaction to external forces.
Indeed every stage of the data-processing pipeline during an image-guided intervention requires a model, including the very initial stage of image formation on a particular imaging modality [17]. The knowledge of the techniques used in MRI or ultrasound image formation, for instance, helps us interpret the images correctly, as well as identify associated artifacts. Similarly, fluoroscopy images can be corrected if a model for the inherent pincushion distortion of a particular fluoroscopy imaging device is known a priori.
Model generation alone, nonetheless, is often used to refer to anatomical modeling of specific patients. In this respect, a very important aspect of model building is the proper representation of the anatomy based on geometric primitives. For this purpose, there exist some simpler techniques that rely on a limited collection of idealized geometric shapes, for instance, cylinders for vessels, ellipsoids for the prostate, and discs for the cornea. These models can help in both segmentation and registration tasks. Such models are, however, only effective when the corresponding anatomy can actually be approximated reasonably by such shapes. More complex organs or simulation scenarios call for more sophisticated models involving mesh representations, which consist of simple geometric primitives such as triangles for describing surfaces or tetrahedra or hexahedra for volumes. These meshes are often generated from the segmentation of the corresponding anatomy in preoperative images. One main concern in this process, however, is the quality of generated meshes (i.e., the distortion of the individual geometric primitives), which may indeed strongly influence the efficiency (or even the feasibility) of numerical procedures used for any subsequent simulation. Figure 4.4 shows a tetrahedral mesh of the pelvic region to be used in the simulation of deformation induced by a rectal ultrasound transducer probe and inserted needles during prostate brachytherapy, which is a treatment for cancer by implanting radioactive seeds using needles [18].
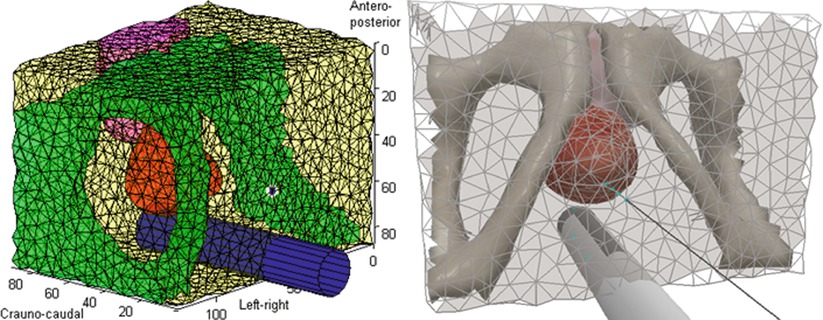

Fig. 4.4
A tetrahedral mesh describing the pelvic region used for simulating tissue deformation induced by the rectal probe and the needle during prostate brachytherapy. The computational model of tissue and probe is depicted on the left, while a realistic rendering used in a training simulation is shown on the right (© [2011] IEEE Reprinted, with permission, from Goksel et al. [18])
Visualization
Traditionally, radiological data are presented to clinicians as 2D images printed on a film and viewed on light boxes. This procedure, even today, offers certain advantages due to the high-resolution and superior dynamic range of analog films. Nevertheless, even for 2D images, digital representation on a computer screen is becoming the standard procedure due to the use of digital sensors in the imaging itself and the versatility of computer tools to exchange and investigate images in networked computer environments.
The broad availability of 3-dimensional (3D) imaging devices and corresponding volumetric anatomical models is another important driving force for the development of computer-based visualization tools. These provide a better understanding of anatomical relations in 3D space, by offering for instance photorealistic rendering of organs based on radiological images. Such visualization techniques are motivated and driven by the need to adequately present the ever-increasing amount of medical data to clinicians in a natural and convenient manner.
The visualization process involves both the rendering of the acquired data and the subsequent output of the resulting images on a suitable display device. The former is mainly a software component, whereas the latter depends on the capabilities of the particular hardware.
For decades, classical 2D screens have been, and still are, the common display hardware in most situations, even though the underlying technology has gone through a major evolution, from cathode-ray tubes to devices using liquid-crystals, plasma technology, or light-emitting diodes. In the meantime 3D display solutions have also been explored [19, 20]. Autostereoscopy, in which special screen (coating) is used to direct different images to each eye, and also 3D displays that utilize special glasses are some examples of recent advances that enable the visualization of sense of depth. Such displays work on the principle that 3D perception is generated in the brain from the two slightly different views of the scene obtained by each eye. Head-mounted displays are used to render scenes according to the head position and direction of the gaze, and as such are of major interest in augmented reality systems in medicine. The stereo vision of the da Vinci system (Intuitive Surgical, Sunnyvale, CA) [21], a surgical robot gaining popularity in laparoscopic procedures, has such a closed-circuit system in which the surgeon at a console can see the 3D anatomy inside the patient acquired by a stereoscopic camera at the tip of a laparoscopic tool. We are currently witnessing how this initially rather expensive technology is now steadily penetrating consumer electronics and meanwhile becoming available on much more affordable, lower-end computer graphic devices.
Rendering refers to the process of image generation from computational models and imaging data. It relies on a simplified – yet sometimes quite involved – model of image formation, often imitating some aspect of the interaction between light and matter, such as reflection or absorption. Ultimately, rendering involves the projection of a scene, such as medical imaging data, onto a plane, in the direction in which the viewer is observing that scene. The projection method is chosen depending on the nature of data to be visualized, such as the triangulated surface of a deformable model or the 3D information contained in a CT image; and accordingly is called surface rendering or volume rendering, respectively. Whether it be a medical tool in the scene or an anatomical organ, surface rendering requires a model of the outer surface of that object. For the anatomy this requires a segmentation step in order to extract that boundary. Once these surface models are known, then well-established methods from computer graphics can be used for rendering [22].
In contrast, volume rendering is a challenge often unique to medical imaging, where data may, and often do, carry information at each and every 3D spatial location (voxel) within a region of interest. Because most objects we encounter in our daily life are opaque, their visual appearance is determined by their surfaces. Nevertheless, objects can also be transparent for visible light, which can penetrate their interior. Volume rendering explores this analogy, casting rays into an image volume, interacting with each voxel on its path of penetration. Different optical properties (like reflectivity, absorptivity, or color) are assigned to each individual voxel, based on the actually measured image data (e.g., intensity) or derived properties (e.g., local direction and prominence of edges assumed to represent anatomical boundaries). The mapping between measured data and optical properties of a voxel is of decisive importance for the quality and meaningfulness of the resulting images and is usually defined in a more or less heuristic fashion. There are, however, a few standard selections which are broadly used in 3D-rendering systems. For instance, two common methods are ray casting, in which each voxel property, e.g., intensity, is integrated along a ray path to project it from a camera viewpoint (imitating X-ray penetration of tissue in a highly simplified fashion), and maximum intensity projection, in which only the maximum intensity along such a path is selected (which has no physical analogy). These inherently costly operations, in terms of memory and computation, traditionally required offline processing and/or limiting the quality of the resulting images. However, on recent computer graphics devices, these and other volume-rendering techniques can be performed at high quality with real-time (or near real-time) refresh rates using hardware acceleration. High refresh rates are essential in interactive volume rendering, because it is often difficult to perceive the depth and to resolve occlusions from merely a single static projection, necessitating the volume to be rotated by the viewer. Such animated, multiple projections from different points of views additionally facilitate the perception of 3D volumetric information [23] presents a detailed review of volume-rendering techniques.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree