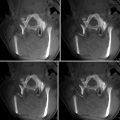
Fig. 36.1
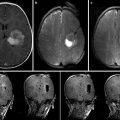
Navigation screenshot. (a) Microscope view after tumor removal with outlined segmented objects. (b–d) T1-weighted MRI images, axial, sagittal, coronal view; objects colored in yellow, tumor; green, language fibers; blue, corticospinal tract; orange, Broca’s area
A 47-year-old patient presented after a first seizure, the cerebral MRI scan showing a left temporal lesion. The neurological examination revealed no further deficits. The patient underwent microsurgical tumor resection, with functional navigation guidance. FMRI was preoperatively performed for Broca’s and Wernicke’s areas, used as seed regions for a DTI-based fiber tracking for language-related pathways. The pyramidal tracts were also displayed in the navigation. An early postoperative MRI proved gross-total tumor resection. The histopathological analysis revealed an oligoastrocytoma (WHO II). Postoperatively, the patient developed a moderate sensorimotor aphasia, which finally completely resolved in a follow-up examination after 6 months.
Magnetic Resonance Spectroscopic Imaging (MRSI)
Multivoxel MRSI or chemical shift imaging (CSI) is based on the interaction of a molecule’s atomic nucleus with the surrounding magnetic field and identifies certain chemical substrates as well as the inter- and intramolecular interactions. MRSI thereby delivers energy spectra displaying the spatial distribution of the nuclear spins while using magnetic field gradients, visualizing shape and spin density of the examined probe. The resulting spectra are characteristic for the chemical binding activity of the atomic nucleus.
For hydrogen nuclear magnetic resonance imaging (H-NMR), the neutral point of the MR spectrum on ppm scale (parts per million) is defined at substrate tetramethylsilane (TMS) and used as reference point for other substrates. This normalization allows evaluation independent of the magnetic field strength. Analysis and evaluation is then based on differences in resonance frequency (chemical shift), being specific for certain metabolites, and aims at their detection and measurement. Finally, the spectra are post-processed and can be evaluated quantitatively.
Brain parenchyma generally obtains spectra for choline (Cho-peak at 3.2 ppm), creatine/phosphocreatine (Cr-peak at 3.0 ppm), and N-acetylaspartate (NAA-peak at 2.0 ppm). Cho is a marker for the integrity of the cell membrane, Cr for intact cell metabolism, and NAA for the integrity of neurons. The spectrum changes characteristically in cases of brain pathologies such as neoplastic or ischemic zones.
In gliomas there is a shift with a decreased NAA-peak and an increased Cho-peak [29, 30]. More precisely, we can find the following concentrations or concentration ratios for neuroepithelial tumors [31]:
Low-grade gliomas: NAA/Cr ↓, Cho/Cr ↑, Cho/NAA ↑
High-grade gliomas: NAA/Cr ↓↓, Cho/Cr ↑↑, Cho/NAA ↑↑, lactate ↑
MRSI has proven to be a dependable preoperative diagnostic tool in cases where the diagnosis is uncertain, as it offers additional information on lesions, which are difficult to distinguish by common MRI sequences, for example, low-grade glioma vs. ischemic lesion vs. certain inflammatory lesions or demyelinating foci (need a reference here). Furthermore the integration of metabolic maps (see Fig. 36.2) into navigation allows segmentation of the tumor’s most metabolically active parts, influencing the surgical strategy or sampling site for biopsy. This improvement of diagnostic yield in brain biopsy was demonstrated by Hall et al. [32], who evaluated a cohort of patients who underwent tumor biopsy with preoperative turbo spectroscopic imaging (TSI), single-voxel spectroscopy (SVS), and real-time needle placement. The findings of SVS correlated with the pathology in all cases; the TSI spectra correlated well with the permanent pathologic examination in 76 %. Martin et al. demonstrated that coupling of TSI targeting with conventional imaging and intraoperative confirmation of needle placement results in 100 % diagnostic success rate [33].
The integration of metabolic maps into the navigation data set enables a spatial correlation of metabolic data and histopathological findings. A study correlating the histopathological results of low-grade gliomas with proton MRS suggested that absolute NAA values are more significant than Cho in the detection of cell infiltration [34]. Furthermore, Stadlbauer et al. showed advantages in glioma border delineation due to high spatial resolution 1H-MRS (voxel size 0.45 cm [3]) combined with a region growing algorithm. Histological findings showed tumor infiltration ranging from 4 to 17 % in areas differentiated from normal brain parenchyma in 1H-MRS only [35].
Single Photon Emission Computed Tomography (SPECT) and Positron Emission Tomography (PET)
SPECT and PET are both based on the application of substances labeled with radioisotopes. Like MRS, they provide more than mere anatomical information, being part of the so-called molecular imaging methods.
SPECT uses various radioisotopes that emit one or more gamma quants, recorded by one or more rotating scintillation detectors (gamma cameras) with connected upstream collimator. Today, the most frequently used scintillator is sodium iodide. The scintillator is followed by a photomultiplier. Here, the photoelectric effect produces electrons which are afterwards transformed in a voltage pulse.
Based on the applied chemical substance and its linkage to the receptor, the characteristic properties of the tissue are imaged. 201-TI or 123-IMT (l-3-123I-alpha-methyltyrosine) SPECT studies have shown an increased uptake in gliomas compared to the surrounding tissue (need a ref here).
PET records high energy gamma rays, emitted by the examined object. Natural molecules are marked with short-living positron emitters (emission energy: several 100 keV to MeV). Due to annihilation of a nuclear positron with an electron, two 511 keV gamma quants spin apart at a 180° angle. These two annihilation quants are registered by ring-shaped detectors arranged around the object [36]. 18F-fluoro-ethyltyrosin (FET)-PET (an amino acid analogue) (see Fig. 36.2) or 18F-fluordesoxyglucose (FDG)-PET (a glucose analogue) can be used for the detection of neuroepithelial tumors. Due to an increased glucose and amino acid metabolism, uptake in tumors is higher compared to the surrounding normal brain.
Stadlbauer et al. evaluated the correlation of tumor invasion into the pyramidal tract in glioma patients with sensorimotor deficits by correlation of 18F-PET and DTI in 2009. They showed an association between preoperative sensorimotor deficits, increased uptake of 18F-FET, and decreased FA ratio in the pyramidal tract, furthermore a correlation between tumor invasion and 18F-FET uptake [37].
Clinical Case (Fig. 36.2)
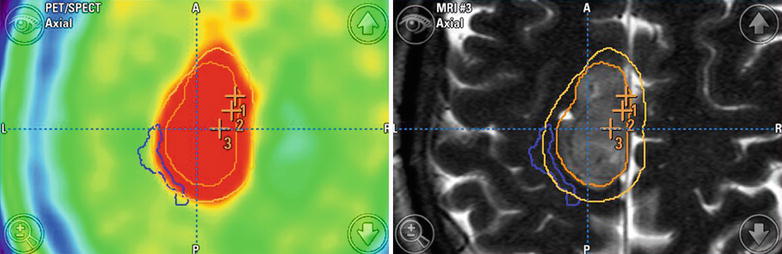
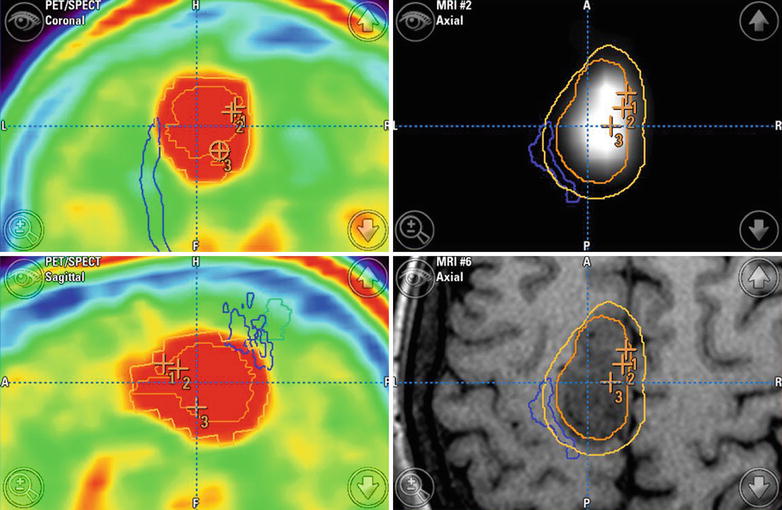
Fig. 36.2
Navigation screenshot including DTI tractography (corticospinal tract: blue), MRSI (right middle image, segmented in dark orange), PET (left-side images), and tumor outlines (light orange, segmentation performed in T2-weighted image data set). The points numbered 1 and 2 mark positions of biopsy samples
A 50-year-old female patient was admitted to our hospital after stereotactic biopsy of a left-sided precentral lesion. Histopathology had revealed an anaplastic astrocytoma (WHO grade III). Due to tumor progression on postoperative MRI, we performed reoperation after integration of DTI tractography, MRS, and PET in the navigation with iMRI resection control. This revealed a small band of residual tumor adjacent to the motor cortex and pyramidal tract. In this way, surgery was not continued after iMRI resulting in a final tumor volume reduction from 12.7 to 2.2 ml. Immediately after surgery there was a new right-sided paresis, which completely resolved at the time of discharge.
Navigation Accuracy
High navigation accuracy is essential during the whole process of surgery, especially in cases of lesions near eloquent cortical areas or major white matter tracts. In addition to MR-image quality and technical accuracy of the navigation system, the two major problems impeding navigation accuracy are the registration process and brain deformations during surgery, the so-called brain-shift problem.
Registration Process
The initial patient registration process is prone to errors. The most commonly used registration strategy is based on skin-adhesive markers placed on the patient’s head, called fiducials, which can be detected in the structural MRI data sets. By defining their position, virtual and physical space can be correlated to create a registration coordinate system. This is called “marker-fit registration” [38]. The “surface-fit technique” uses the contour of skull and face for the referencing process [39]. User-dependent errors have been overcome by the development of semiautomatic and automatic registration processes. With the semiautomatic registration process, the markers in image space are detected automatically, whereas the rigid registration process itself is performed manually [40]. However, with intraoperative MRI automatic registration allows user-independent registration of patient space and image space [41]. The registration matrix as a fixed constellation of fiducials is attached to the upper part of the head coil so that the fiducials are automatically imaged. After scanning, the acquired images are transferred to the navigation system.
The infrared navigation camera identifies the reflective marker structure of the registration matrix and the reference array and determines the spatial relation between them. An automatic marker detection algorithm detects the fiducials from the registration matrix in the image data set. The information about the exact fiducial arrangement is used as input for the paired point matching algorithm. A transformation matrix is calculated, combining the following information:
1.
The position of the registration matrix in relation to the reference array
2.
The relation of the detected fiducials to the fiducials of the registration matrix
3.
The relation of the fiducials of the registration matrix to the spatial position of the matrix itself
In this way, the reference array is directly related with the acquired images so that a relation between image space and physical space is defined. An additional skin fiducial attached to the patient’s head is used to document the target registration error (Fig. 36.3).
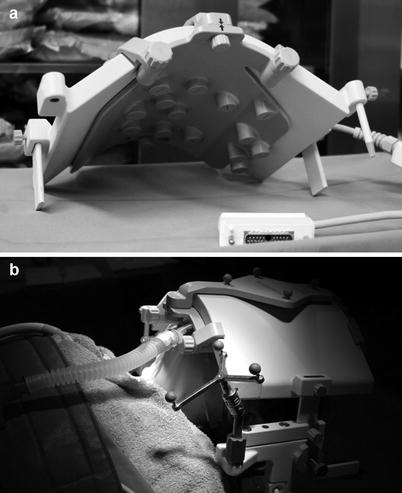

Fig. 36.3
(a) Upper part of MRI head coil with equipped registration matrix as fixed constellation of fiducials. (b) Setting before iMRI, showing the reflective marker structures reference array and registration matrix at the top of the head coil
Brain Shift
Another factor influencing navigation accuracy, especially as surgery progresses, is the so-called brain-shift problem. This describes intraoperative deformations of the brain mainly due to drainage of cerebrospinal fluid, tumor resection, use of retractors, or brain swelling [42, 43]. Thus, navigation systems relying on the preoperative image data sets have decreasing accuracy as the surgery proceeds. Brain shift can be compensated for by intraoperative imaging followed by updating the navigation with the new intraoperative image data. With the intraoperative images the actual physical reality can be reconstructed virtually, including the effects of brain shift and EOR [19, 44–46].
Intraoperative MRI (IMRI)
Intraoperative imaging methods have attracted the interest of neurosurgeons as objective methods to determine the extent of tumor resection. As the surgeon’s subjective estimation is not always entirely reliable, intraoperative imaging enables the surgeon to remove missed tumor remnant during the same operation. Among the intraoperative imaging modalities, computed tomography, ultrasound, and 5-ALA, MRI offers the highest resolution for the detection of even small tumor remnants. Furthermore it allows updating the navigation, which is described in the following. In this way iMRI serves as immediate quality control and contributes to higher rates of EOR and gross-total resection (GTR) without increased morbidity.
In the mid-1990s, the development of open MRI scanners allowed integration of this technique into operating theaters. From initially established low-field scanners, the development to intraoperative high-field MRI was initiated by Hall et al. [47] and Sutherland et al. [48], which also facilitates the integration of advanced MRI sequences like DTI.
With the help of intraoperative MRI acquisition, brain shift can be compensated and used to update the navigation data set, by registration of the intraoperative image data, though this may be a cumbersome process. Previously, bone fiducials were placed around the craniotomy opening and then used for reregistration. This method was time consuming and of low accuracy. Later, high-field scanners and microscope-based navigation led to further registration and update facilities:
1.
The same calibrated registration matrix, which has been used to register the preoperative images, can be attached to the upper part of the sterile head coil and tracked by the navigation system after iMRI. The navigation system automatically detects the markers by tracking the reference star, connected to the head clamp. Thus, the spatial relation to the registration matrix is defined [41, 49].
2.
Alternatively, updating the navigation without intraoperative reregistration is possible. After iMRI, the image data set is transferred to the planning computer of the navigation system and furthermore fused by a user-dependant alignment followed by a rigid registration algorithm [50, 51]. This process is followed by a verification process using the so-called “spyglass” feature. This data is then sent to the navigation computer and the navigation software restarted. The user has to accept the preoperative registration, after which the initial patient registration is restored so that the navigation coordinate system of the preoperative images can be displayed beside each other and contours can be displayed in all images.
Outlook
IMRI followed by an update of navigation is routinely used with anatomical MR images, thus compensating for the effects of brain shift. However, an intraoperative update of functional data, e.g., fMRI and DTI, is still not commonly used due to time constraints. FMRI during the intraoperative imaging requires awake surgery for the performance of language fMRI paradigms. The lack of functional update might be overcome with either nonlinear registration techniques or sophisticated techniques from pattern recognition, allowing the matching of preoperative data sets including functional data with the intraoperative MR images [52, 53].
For compensation of brain shift, iMRI with intraoperative update of the navigation is the gold standard. However, intraoperative 3D ultrasound combined with nonlinear registration to preoperative MRI data sets is an alternative approach for brain-shift compensation [53, 54]. With mathematical models the MRI images are transformed, describing the deformations of the brain during surgery [55–57]. In addition to these models, simulating the effects of brain shift, there are further mathematical models describing the actual intraoperative 3D situation in combination with sparse data [57, 58].
For the various methods to display functional data, future work with ongoing technical advances will provide better image resolution and thus more reliable information.
Especially for fiber tractography of white matter tracts, there are promising new developments.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree