3D TEE Imaging
INTRODUCTION
Real-time (RT) three-dimensional (3D) TEE is being increasingly used in the perioperative period to assess cardiac anatomy and function. Ease of acquisition with rapid online display of detailed dynamic 3D images has overcome some of the early limitations associated with 3D echocardiography. A basic understanding of evolving 3D technology enables the echocardiographer to master the new skills necessary to acquire, manipulate, and interpret 3D datasets (1–3). Cardiac structures can be shown from any perspective including a virtual surgical orientation. This single display of a dynamic image is an invaluable visual aid to the echocardiographer and surgeon to better communicate and understand individual patient anatomy. In addition, exportation of 3D datasets to analytical software permits prompt off-line reconstruction of 3D models to accurately quantify cardiac anatomy and ventricular function. This chapter presents key features of the emerging 3D technology used in echocardiography and reviews the current applications of RT 3D TEE.
3D TECHNOLOGY
A fundamental difference between two-dimensional (2D) and 3D echocardiographic imaging is how the image is acquired and displayed. Volume scanning is used in RT 3D echocardiography as compared with the standard sector planes in 2D (4). This requires the use of special ultrasound probes to acquire raw data and integrated ultrasound machine software to process 3D datasets.
TEE Probes
The modern adult 2D TEE multiplane transducer comprises a linear phased array of 64 to 128 piezoelectric crystals arranged to cover a circular transducer face. Sequential activation of the crystals generate a 2D, sector-shaped, flat, ultrasound plane that can be mechanically or electronically rotated in 1-degree increment through 180° to scan a conical-shaped area. Three-dimensional images can be created using a standard 2D multiplane TEE probe but this is time-consuming and requires off-line reconstruction.
RT 3D echocardiography uses transthoracic echocardiography (TTE) and transesophageal echocardiography probes with special matrix array transducers. In a similarly sized 5 to 7 MHz 3D TEE probe (X7-2t, Philips Healthcare, Andover, MA), extreme miniaturization assembles 2,500 piezoelectric crystals arranged in 50 rows by 50 columns to cover the entire square transducer face. Each crystal can be independently activated (fully sampled), focused, and steered. This generates an ultrasound beam that can be steered in the azimuthal (x-y) and the elevational plane (x-z) to cover a 3D pyramidal scanning volume (Fig. 21.1). In addition, these probes can also perform all the standard 2D functions including M-mode, Doppler (spectral, color, and tissue) modes and are also able to simultaneously display two independent 2D scanning planes (xPlane mode).
3D Imaging
Creation of a 3D ultrasound image of the heart involves four basic steps: Data acquisition, data storage, data processing, and image display (Fig. 21.1A).
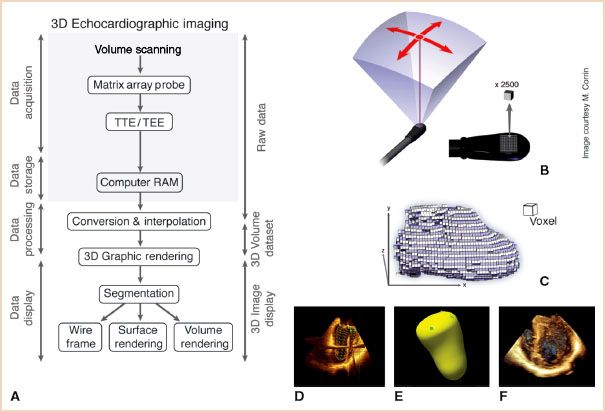
FIGURE 21.1 3D imaging. A: 3D echocardiographic imaging includes four stages: Data acquisition, data storage, data processing, and data display. B: A matrix array probe comprising 2,500 piezoelectric crystals is used to volume scan and acquire raw 3D data. C: Data processing involves creation of a 3D dataset comprising voxels through conversion (white) and interpolation (purple). D–F: 3D data can be displayed as (D) wireframe, (E) surface, or (F) volume rendered as shown here for the left ventricle. Images (B) and (C) used with permission of M. Corrin, (D–F) from: Vegas A, Meineri M. Three-dimensional transesophageal echocardiography is a major advance for intraoperative clinical management of patients undergoing cardiac surgery: A core review. Anesth Analg. 2010;110:1548–1573, with permission. Video 21.1.
Data Acquisition
Initial 3D data acquisition obtains echocardiographic characteristics of many single points within a volume of tissue. The matrix array probe is held immobile while the transducer automatically steers the ultrasound beam to scan a pyramidal volume of tissue in three dimensions.
Data Storage
Data storage maintains data flow from the initial raw data acquisition to the next step of data processing. During the volume scanning technique, data is streamed through the onboard computer, allowing for immediate concurrent data storage and processing (online) within the ultrasound machine. Off-line exportation and reconstruction of the 3D dataset is no longer required.
Data Processing
Data processing consists of two integrated steps: Conversion and interpolation that transform the scanned raw data for a specific volume into a 3D dataset used to create a 3D object (Fig. 21.1C).
Conversion positions and assigns x-y-z coordinates (Cartesian volume) to each of the scanned raw data points identifying their echogenic characteristics with a known position in space. Interpolation fills the gaps between all the known points in space to generate a 3D dataset comprising voxels or volume elements. A voxel is a (vo)lume of pi(xel)s that encrypts the physical characteristics and location of the smallest cube in a dataset, which is used for 3D display. Resolution of the 3D image depends on voxel size. Larger voxels resulting in poorer resolution are generated when raw data is available for fewer points in the scanned volume and interpolation has to fill wider gaps.
3D Display
The process of making a 3D dataset visible is termed 3D display which can be either as multiple 2D image planes or a 3D graphic reproduction.
Graphic rendering is a computer graphics process that creates a 3D object or image. Segmentation is the first step in graphic rendering during which the object to be rendered is identified and separated from surrounding structures in the 3D dataset. This consists of differentiating cardiac tissue from blood, pericardial fluid, and air which is facilitated by their dissimilar ability to reflect ultrasound. It is achieved by setting the threshold of echo density for blood thus excluding any point with a lower echo density from further processing. This step delineates the 3D surfaces of cardiac tissue.
Following segmentation, the 3D dataset undergoes one of the three increasingly complex graphic rendering techniques: Wireframe, surface, or volume rendering (Fig. 21.1D–F) to display the 3D object.
Wireframe rendering is the simplest and fastest way of creating a visible 3D object. A series of equidistant points on the surface of a 3D object are defined and connected with lines (wires) to form a mesh of small polygonal tiles. Smoothing algorithms can refine the narrow angles and make the object appear more real. In clinical practice this technique is used for structures with relatively flat surfaces such as the left ventricular (LV) and the atrial cavities (Fig. 21.1D).
Surface rendering extends the wireframe technique by defining more points on the surface of a 3D object making the lines joining them invisible. It generates 3D objects with detailed solid surfaces but an empty hollow inner core (Fig. 21.1E). This makes morphologic assessment of the corresponding anatomical structure (e.g., cardiac valves) and determination of cavity volume (e.g., LV volume) feasible.
Volume rendering retains all the 3D data and displays a 3D object with full details of the surface and inner structure. This enables the potential display of every voxel of the 3D dataset permitting a “virtual dissection” of the 3D object (Fig. 21.1F).
Despite being composed of voxels, these 3D objects are seen on the screen as pixels of a 2D image. Perspective, light casting, and depth color coding are used to give a visual sense of depth and reality. Any rendered 3D object can be freely rotated in the display and shown from any orientation. Furthermore the 3D datasets can be displayed as a static or a moving object. A moving (dynamic) 3D object is often referred to as four dimensional (4D), with time considered as the fourth dimension.
3D IMAGE ACQUISITION
The acquisition of 3D-TEE images can occur instantaneously either in (a) RT (live) or (b) gated that is timed to the electrocardiogram (ECG) over multiple heartbeats. Currently the term “real time” refers to any 3D image that changes on the display with probe movement. Gated images are created as a loop by stitching together subvolumes acquired from consecutive heartbeats. Over a fixed number of cardiac cycles, the same portion of the ECG wave (usually the R wave) triggers the recording of each subvolume and allows rapid, within seconds, data reconstruction synchronous with the ECG. It works best for a regular rhythm. Arrhythmias, electrocautery artifacts, and probe movement will affect the ability to seamlessly stitch together the subvolumes. A demarcation line, termed a stitch artifact, appears between the subvolumes distorting the anatomical structures potentially affecting analysis.
All forms of ultrasound imaging are limited by the speed of sound (1,500 m/s). RT 3D TEE has a complex interdependence of temporal resolution, sector size, and image resolution. In 3D echocardiography, temporal resolution can be described in terms of volume rate or frame rate (FR) as shown on the display. There is insufficient time for sound to traverse large tissue volumes in the RT scanning modes. For instance, at a depth of 12 cm, scanning in 2D (90 scan lines) requires 14.8 milliseconds giving a 62 Hz FR with 3D (2,400 scan lines), it requires 396 milliseconds resulting in a 1 to 11 Hz FR. Current-gated acquisition and evolving 3D technology is being developed to produce large 3D volumes in RT with good temporal and spatial resolution.
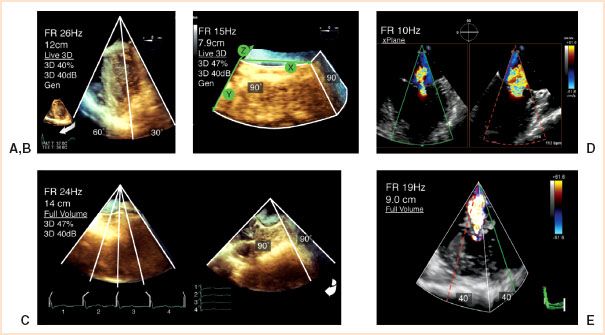
FIGURE 21.2 3D imaging modes. The various 3D imaging modes display assorted volumes of information at different frame rates; (A) Live (up to 30° × 60°) of a rotated midesophageal four-chamber (ME 4-C) view (inset); (B) Zoom (90° × 90°) of an unrotated en face view of the mitral valve; (C) Full volume (90° × 90°) of an ME 4-C view as four segments joined together and rotated. A mitral regurgitation jet is shown in (D) 2D using xPlane and (E) 3D color Doppler Full Volume which has the smallest volume (40° × 40°) and a 19-Hz FR despite containing seven consecutive segments. (Modified from: Vegas A, Meineri M. Three-dimensional transesophageal echocardiography is a major advance for intraoperative clinical management of patients undergoing cardiac surgery: A core review. Anesth Analg. 2010;110:1548–1573, with permission.) Videos 21.2, 21.3, 21.4.
Imaging Modes
Single button activation of specific 3D imaging modes (iE33 machine, Philips Healthcare) for both TEE and TTE matrix array probes include (a) Live, (b) Zoom, (c) Full Volume (FV), and (d) color Doppler FV (Fig. 21.2). Choosing between modes for specific structures is a balance between selecting pyramidal image size, volume or FR, and RT imaging (Table 21.1).
TABLE 21.1 3D Imaging Modes
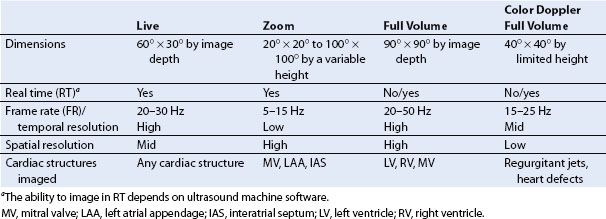
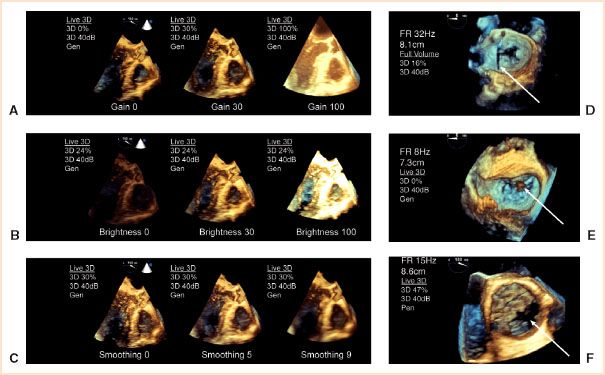
FIGURE 21.3 3D image optimization and artifacts. Postprocessing image optimization of displayed 3D datasets involves adjusting (A) gain, (B) brightness, and (C) smoothing. Artifacts in 3D images include (D) stitch artifact (arrow) between adjacent segments in a full volume en face MV view. E: Overgain appears as brown speckles (arrow) which may be mistaken for a mass in the left atrium in this Zoom en face MV view. F: Dropout from shadowing (arrow) is similar to a 2D image as shown for a calcified bicuspid aortic valve in SAX using live mode. (From: Vegas A, Meineri M, Jerath A. Real-time three-dimensional transesophageal echocardiography. A Step-by-Step Guide. New York, NY: Springer; 2012, with kind permission of Springer Science + Business Media.) Video 21.5.
Depending on the ultrasound machine software, 3D datasets for the same-sized volume in all 3D modes can be acquired and displayed in RT over multiple heartbeats (1,2,4,5) increasing temporal resolution. The acquisition time is longer for more beats. The single beat acquisition has the lowest FR and is useful with arrhythmias as it does not require an ECG.
(a) Live displays a RT narrow angle 3D pyramidal volume with fixed dimensions of 60° × 30° by the same depth of the initial 2D view for one or multiple heartbeats. Cardiac structures are imaged in 3D using the familiar 2D scanning planes. Rotation of this 3D image can be used as a quick test of the general 3D image settings (Figs. 21.2A and 21.3A). The limited sector size necessitates physical probe movement to image entire structures.
(b) Zoom displays a RT magnified subsection of 3D pyramidal volume that can vary in dimensions from 20° × 20° to 100° × 100° by a variable height of the 2D view. The 3D pyramidal volume is centered to a specific region of interest (e.g., the mitral valve [MV]) and kept as small as possible in order to improve the FR and image definition (Fig. 21.2B).
(c) Full Volume is an ECG-gated or RT acquisition of a large 3D pyramidal volume with dimensions up to 90° × 90° by the height of the 2D view. Individual wedge-shaped subvolumes created over consecutive heartbeats are stitched together and synchronized to the same cardiac cycle (Fig. 21.2C). Different FV acquisition options can be chosen to optimize (a) volume size (seven beats over large sector), (b) ECG (four beats over large sector), or (c) FR (seven beats over small sector). A machine preset (auto crop 50%) initially displays only half of the volume-rendered image revealing its inner details (Fig. 21.1F).
(d) Color Doppler FV is a gated or an RT acquisition of a small 3D pyramidal volume with superimposed 3D representation of color Doppler flow. Similar to Zoom, color sectors with an appropriate Nyquist are centered on two orthogonal 2D color Doppler views to represent the 3D volume. Stitching artifacts are common as acquisition involves multiple subvolumes. Given the amount of information (3D volume and 3D flow), the current technology only creates a small (up to 40° × 40°) 3D volume at a low FR <20 Hz (Fig. 21.2D).
POSTPROCESSING
Image Optimization
Important determinants of 3D image quality and size during image acquisition are line density and transducer frequency. Line density is the number of scan lines per volume and affects the sector size and spatial resolution. A low line density results in a larger sector size but poorer spatial resolution. Transducer frequency is determined by the selection of options such as (a) penetration (low frequency), (b) general (intermediate frequency), or (c) resolution (high frequency), and like in 2D imaging, can dramatically affect the image appearance. The quality of the 3D image starts with adjustment of gain and compression to optimize the 2D image. Gain, brightness (increasing whiteness), and smoothing (reducing image coarseness) are adjustable 3D settings in RT or on any stored 3D datasets (Fig. 21.3A–C).
Image Artifacts
As previously described, a stitch artifact occurs between subvolumes in an FV-rendered image (Fig. 21.3D). Excessive gain in a 3D image results in noise that appears as brown speckles and may obscure structures (Fig. 21.3E). Dropout is an absence of echoes and appears black on the display (Fig. 21.3F). It may result from insufficient gain, shadowing, or ultrasound attenuation. Thin (interatrial septum [IAS]) and distant structures (aortic and pulmonic valves) remain difficult to image by 3D TEE. Most artifacts present in a 2D image will also appear in the 3D image.
Cropping and Orientation
Any 3D volume can be cropped, rotated, and displayed from any perspective (Fig. 21.4). It is possible now to present intraoperative RT 3D TEE images in the surgeon’s orientation. The easily recognized aortic valve (AV) is often used as a reference to orient the 3D volume in space. Alternatively, the reference 2D orthogonal planes can be displayed to guide orientation (Fig. 21.4C).
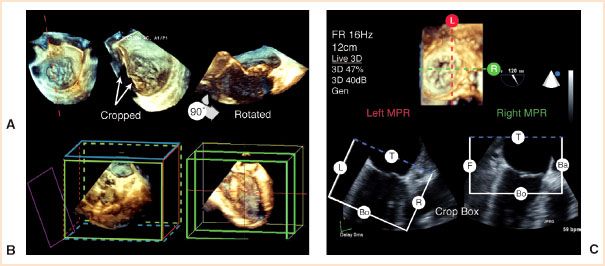
FIGURE 21.4 Cropping and rotation. All 3D datasets can be cropped along any plane and freely rotated to be displayed in any orientation. A: Shown here is a prolapsed P1 scallop of the mitral valve (MV) in an en face view from the left atrium (LA), cropped along the red dotted line and rotated 90° to a sagittal plane. B: Cropping can utilize a crop box to cut along predefined planes or a mobile plane (purple). C: Cropping in real-time positions and adjusts a crop box to a region of interest with simultaneous display of any side of the crop box. Shown here is a 3D en face view of the MV from the LA which corresponds to the top of the crop box with reference 2D multiplanar reconstruction (MPR) images. Ba, back; Bo, bottom; F, front; L, left; R, right; T, top. (From: Vegas A, Meineri M, Jerath A. Real-time three-dimensional transesophageal echocardiography. A Step-by-Step Guide. New York, NY: Springer; 2012, used with kind permission of Springer Science + Business Media.)
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree