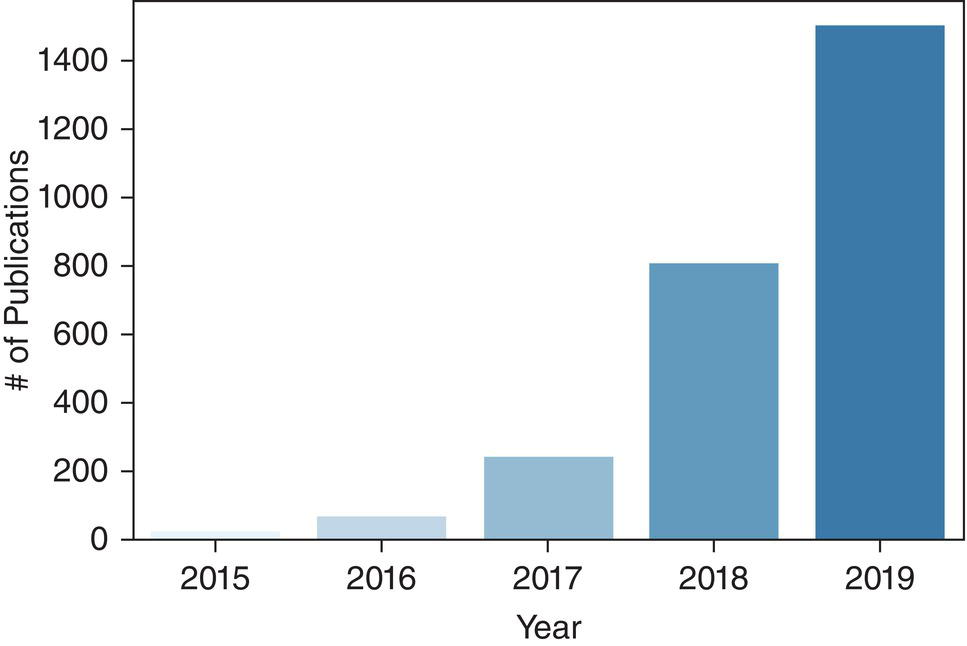
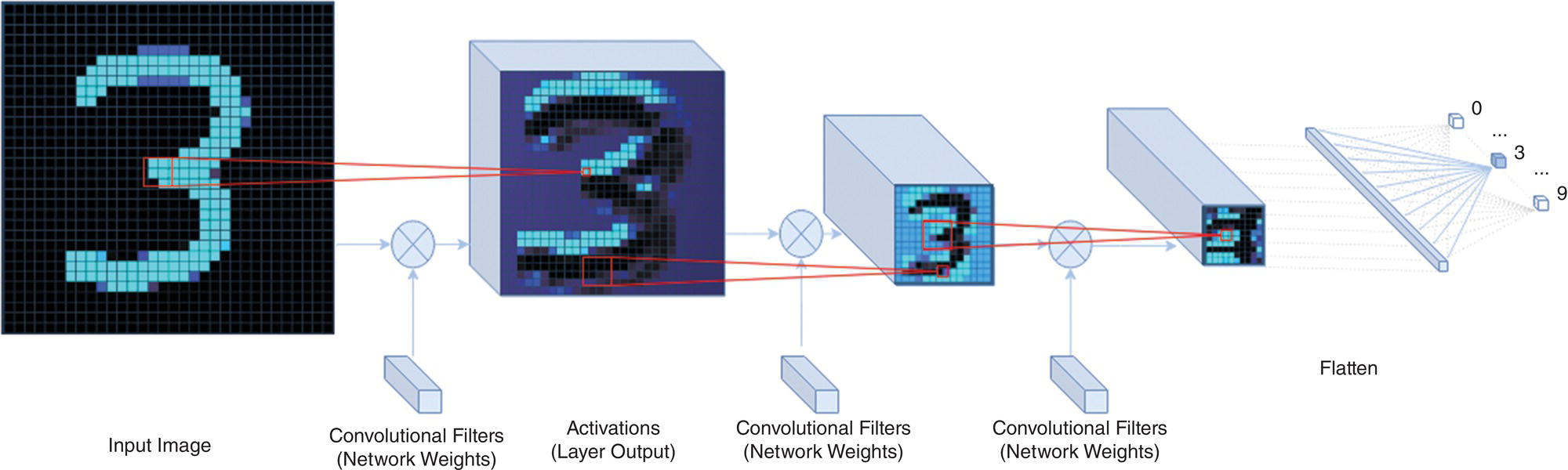
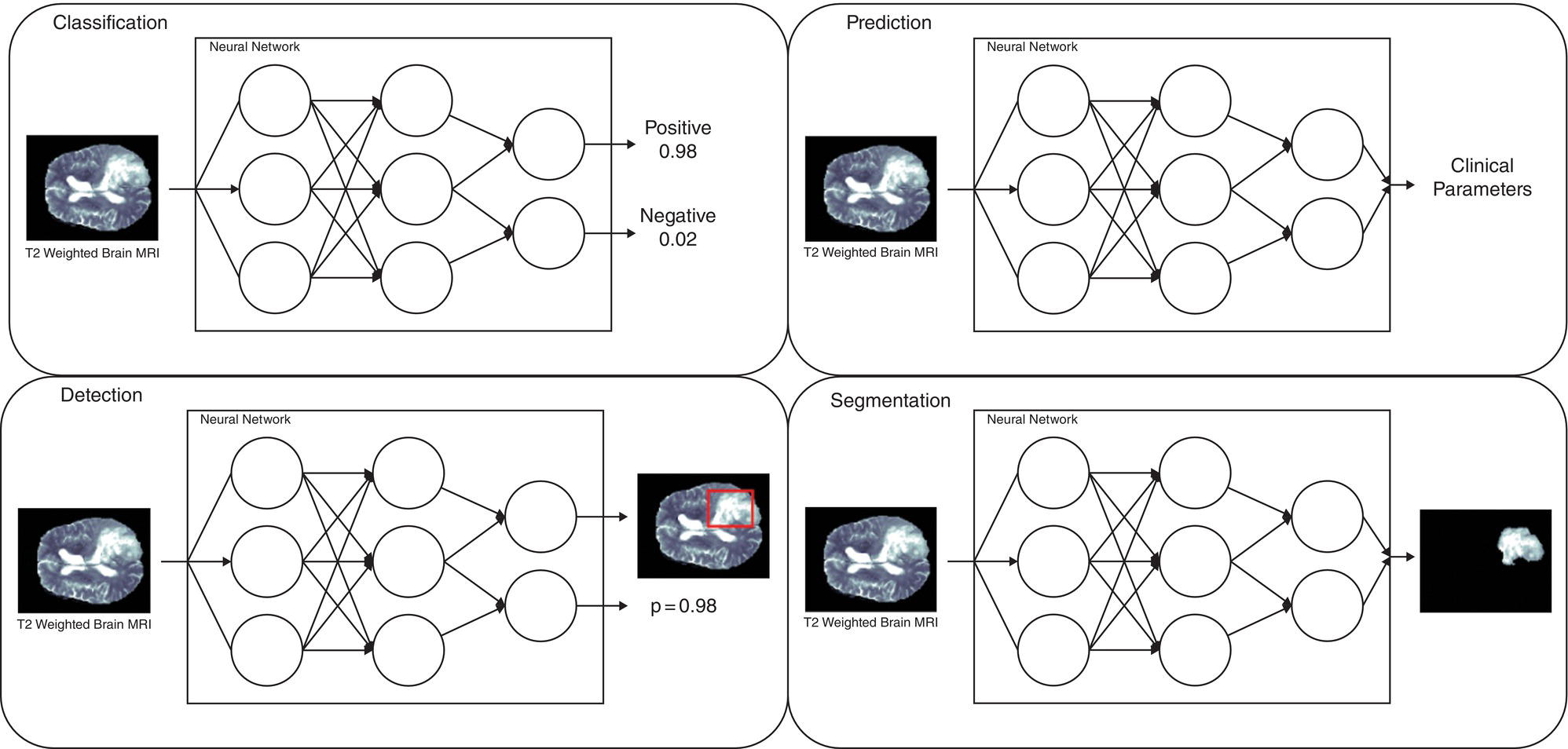
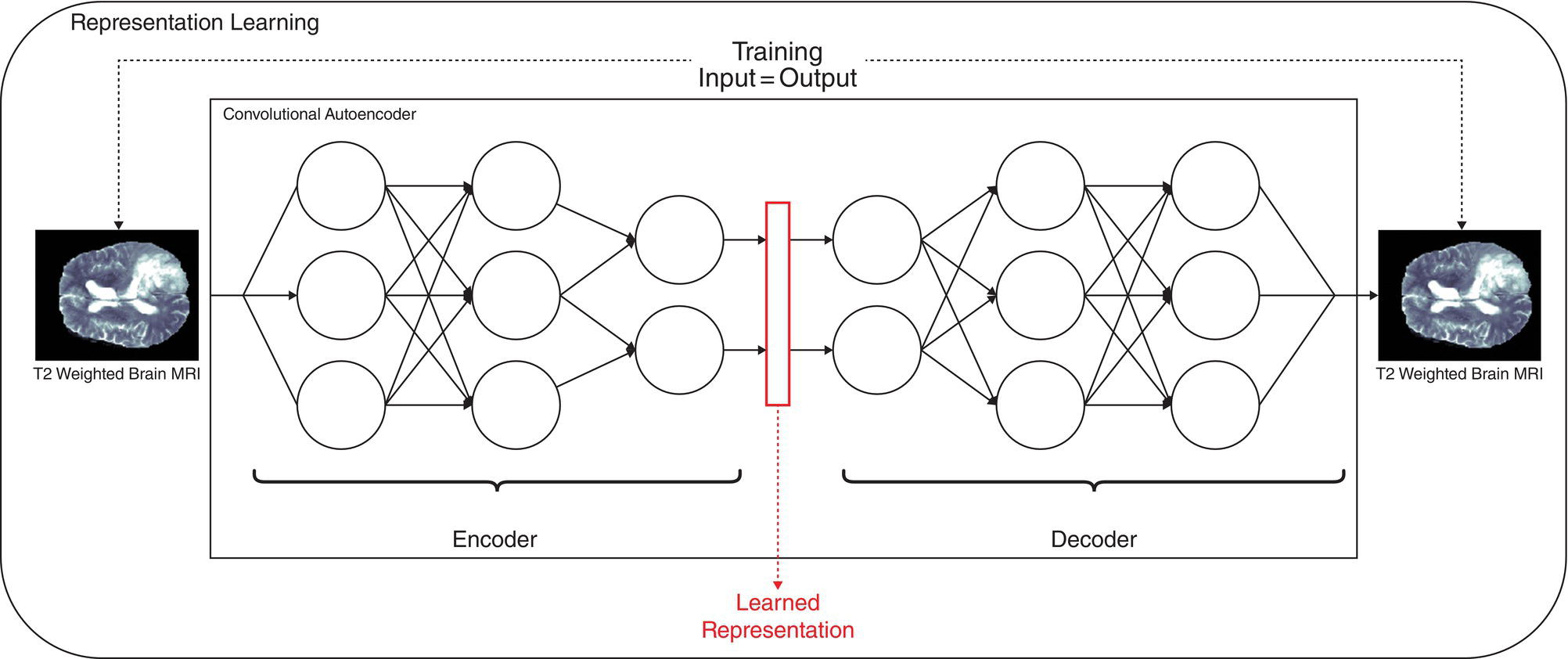
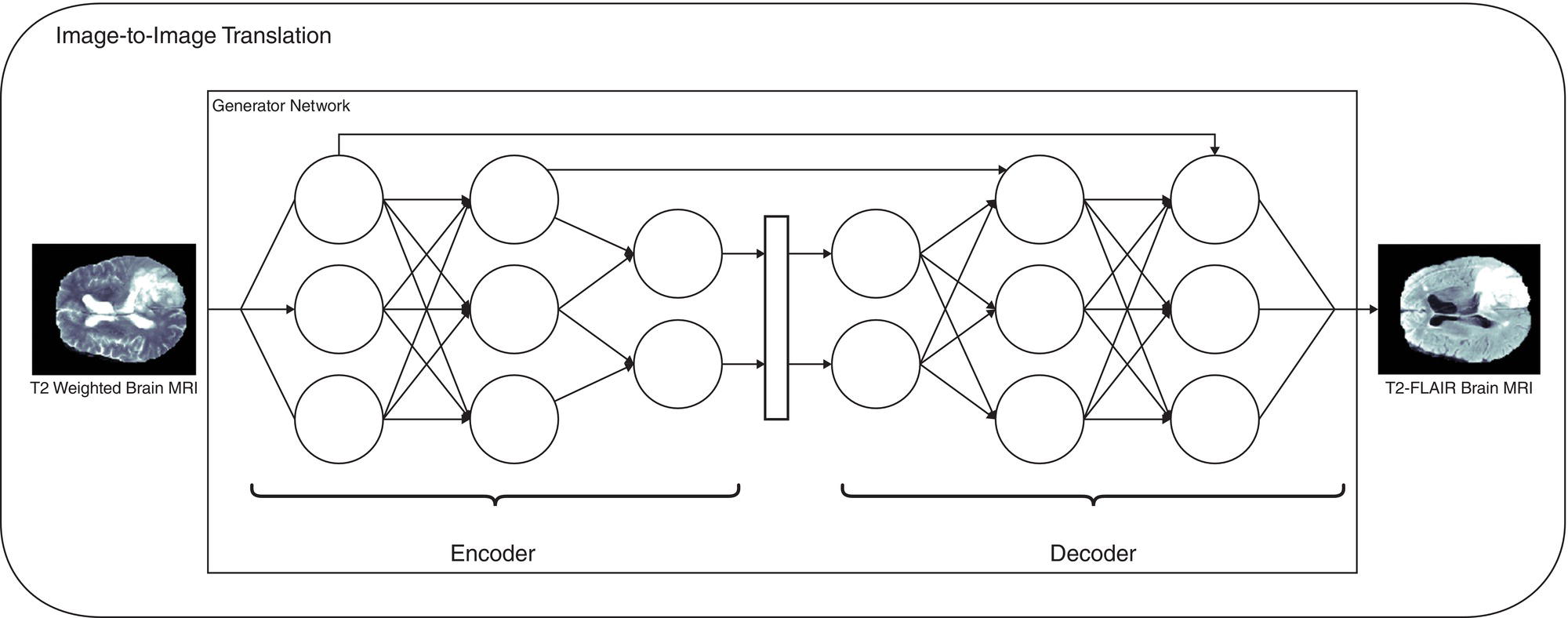
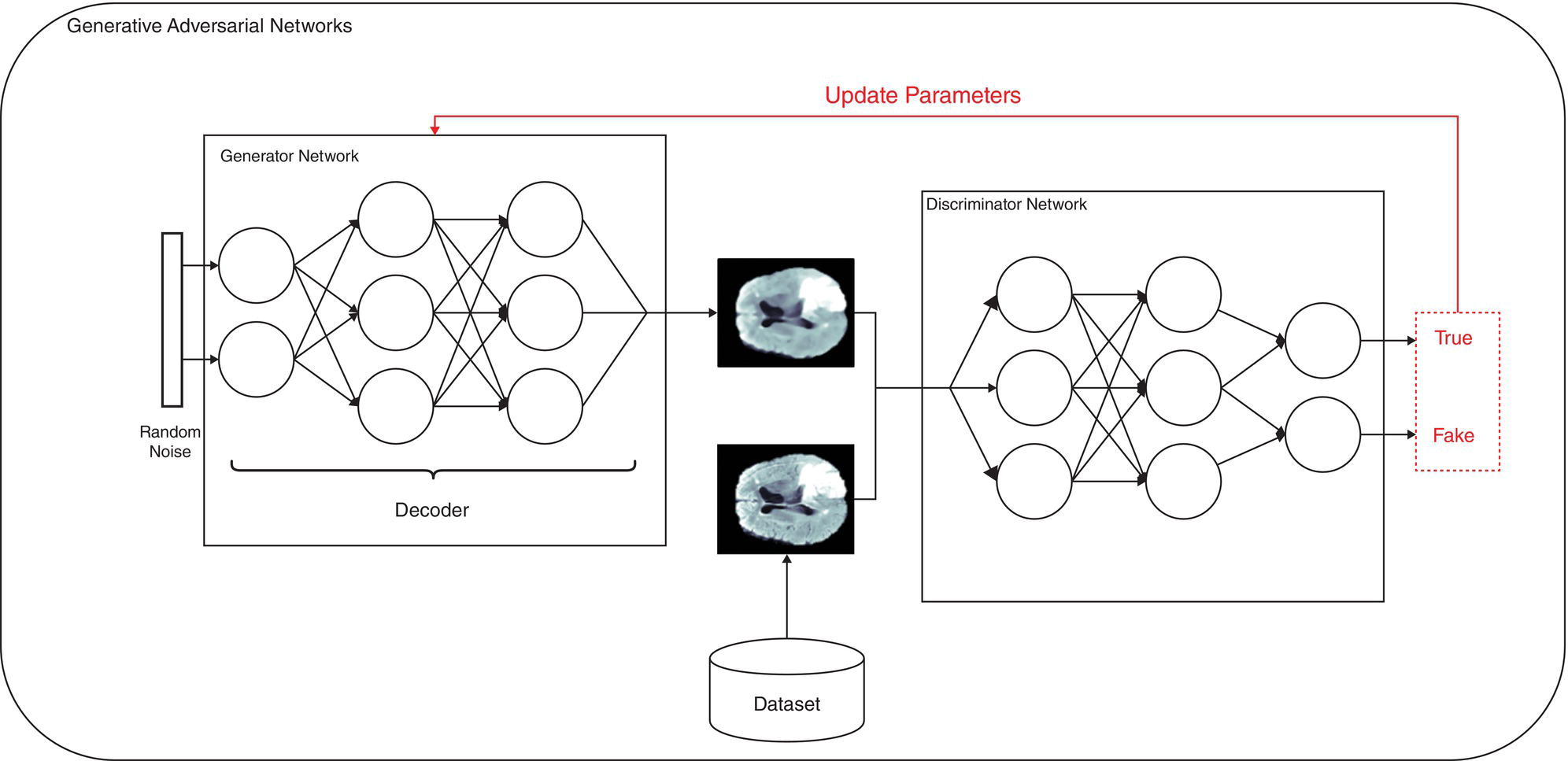
Martina Sollini1,3, Daniele Loiacono2, Daria Volpe1,3, Alessandro Giaj Levra3, Elettra Lomeo3, Edoardo Giacomello2, Margarita Kirienko4, Arturo Chiti1,3, and Pierluca Lanzi2 1 Department of Biomedical Sciences, Humanitas University, Via Rita Levi Montalcini 4, 20090 Pieve Emanuele – Milan, Italy 2 Dipartimento di Elettronica, Informazione e Bioingegneria, Politecnico di Milano, Milano, Italy 3 IRCCS Humanitas Research Hospital, Via Manzoni 56, 20089 Rozzano – Milan, Italy 4 Department of Nuclear Medicine, Istituto Nazionale per lo Studio e la Cura dei Tumori, Milano, Italy Artificial intelligence (AI) research has developed machines able to perform many tasks that normally require human intelligence. Among all AI techniques and methods, machine learning (ML) includes all those approaches that allow computers to learn from data without being explicitly programmed. ML has been extensively applied to medical imaging [1], and the last few years have seen the rise of deep learning (DL), which is an area of ML focused on the application and training of artificial neural networks with a very large number of layers, also called deep neural networks. The major strengths of DL are the capability to extract relevant features from raw data and learn effective data representations, without the need for any human intervention, whereas most ML approaches rely on manually extracted features and human‐designed data representations [2–4]. The introduction of AI algorithms to clinical practice would lead to important benefits, including less time to analyze images (i.e. faster diagnosis), higher expertise and standardization in image interpretation (i.e. accurate description of clinical findings), and a low rate of misdiagnosis (i.e. early and personalized therapeutic approaches). All these benefits would ultimately result in a significant improvement in the patient’s outcome. On the other hand, such algorithms are currently focused on specific tasks, without the possibility of approaching human or disease complexity in a holistic way. Moreover, the dataset used for the development of AI might affect its generalizability (i.e. target population, geographic distribution of endemic diseases). Other general concerns related to AI in the healthcare domain are the accuracy of data used for algorithm development, the size of the training dataset, application to rare conditions or diseases, and legal, ethic, and privacy issues. This chapter aims to provide some insights into AI, including basic knowledge on ML and DL algorithms, challenges, and future directions. Finally, it provides some examples of AI applications in diagnostic imaging. DL emerged as a research area of ML, which is a vast field of AI that includes a variety of methods such as support vector machines (SVMs), decision trees, random forests, clustering algorithms, and neural networks. In particular, DL focuses on methods that stem from the field of neural networks. Indeed, the adjective “deep” refers to the enormous number of artificial neuron layers that are used in these approaches, in contrast with the “shallow” neural networks commonly used in ML. LeCun et al. [5] proposed LeNet as the first successful application of convolutional neural networks (CNNs) in handwritten document recognition, using a seven‐layered network. While CNNs existed for decades, researchers regained interest in neural networks only at the beginning of the 2010s, mainly thanks to the technological advances that allowed graphics processor units (GPUs) to be used to speed up neural network computations and the increased availability of image data. In 2012, Krizhevsky et al. designed AlexNet [6], a CNN inspired by LeNet but much bigger and with many more layers. AlexNet won the ImageNet 2012 competition in which proposed algorithms are asked to classify each image in a dataset among 1000 classes. This result highlighted the effectiveness of CNN in solving image analysis and computer vision tasks and determine the success of DL [7]. The major advantage of DL over other ML approaches is that the deep structure allows more complex and higher‐level patterns to be captured directly from the data. As a notable example, in 2012 Le et al. [8] created a deep neural network capable of learning various concepts from images, such human or cat faces, without the need to know whether each image contained the concept or not. This capability of learning high‐level concepts from the data itself is one of the strengths of DL: while other ML algorithms require an effective feature extraction phase to be successfully applied, DL algorithms can learn an effective data representation automatically. Furthermore, data representations learned from data are generally also useful for related tasks with limited effort [9], whereas feature extraction is usually a problem‐dependent and time‐consuming task. In the last few years, the application of DL to medical imaging has attracted a lot of interest, as proved by the increasing number of papers published in the last 5 years on these topics (see Figure 31.1). On the other hand, despite being very promising, these techniques pose several challenges that are still subject to research, such as the availability of a large amount of labeled and annotated data as well as the limited number of public datasets and models shared by the research community. A key element that has allowed the successful application of DL to images and, more generally, to data with a meaningful spatial distribution, is the introduction of CNN. CNNs are neural networks that apply the convolution operation to extract features from overlapping image patches (see Figure 31.2). This operation in neural network layers relies on the assumption that pixels of an image are more tightly correlated with near pixels than distant ones. This allows the network to learn a high number of spatially small filters (typically 3 × 3 or 5 × 5 pixels) that represent the local features in the input image. In a deep neural network several convolutional layers are stacked one onto another. Accordingly, the first layers of the network can learn low‐level features in the image, such edge orientations, while subsequent layers build their representation based on the previously learned features. An important property of convolutional layers is that they allow translational invariance: thanks to the convolution operation, which applies the same filter on overlapping patches of the input, each layer can detect the same feature independently from its spatial location. Figure 31.1 Number of publications on deep learning applied to medical imaging. Data retrieved from PubMed using the following search string: (“deep learning” OR “deep neural network” OR “convolution neural network”) AND (radiography OR x‐ray OR mammography OR CT OR MRI OR PET OR ultrasound OR therapy OR radiology OR MR OR mammogram OR SPECT). As an example, let look at training CNN to classify photos. The first layers of the network will learn the finest details in an image, such edges, the intermediate layers will combine the edges to recognize patterns as fur, grass or sand, and finally the deeper layers will combine these features to recognize shapes and high‐level concepts like animals, trees, outdoor landscapes, people, etc. DL can be applied to a large variety of tasks. Depending on the data availability and the desired outcome, it is possible to identify different learning approaches: Figure 31.2 Structure of a convolutional neural network for classifying handwritten numbers. Multiple layers of two‐dimensional convolution operations are stacked to extract local features and reduce spatial dimensionality. At each convolutional layer, every image patch is multiplied by a set of filters, which are learned during the training process. In the last layer, a fully connected layer acts as a classifier to produce the final output. The image was generated with an interactive tool for the visualization of convolutional neural networks developed by Harley [10]. Source: Based on Harley [10]. Figure 31.3 An overview of the most common discriminative learning approaches used in deep learning. Figure 31.4 An overview of how deep learning exploits representation learning. Figure 31.5 An overview of an image‐to‐image translation task using a generative model. Figure 31.5 shows how DL is applied to image‐to‐image translation tasks: a generator network, composed by an encoder stage and a decoder stage (as in the autoencoder model described before), is trained to generate images as close as possible to the examples available in the training data. Unfortunately, this training process is complex and often fails to provide the desired results. For this reason, in recent years a novel type of deep neural network has been proposed in the literature: generative adversarial networks (GANs) [13]. This kind of approach has proven to be successful in many computer vision tasks, in particular those related to image generation and translation. The basic idea of GANs (see Figure 31.6) is to train two deep neural networks that work in an adversarial setting. The first is the generator, whose objective is to solve the image‐to‐image translation task as seen before. The second network is the discriminator, or adversarial network, which is a network that takes in input that is either the ground truth samples, from the training dataset, or the samples generated by the generator. The discriminator is trained to classify whether a given sample comes from the dataset or it has been generated, assigning them the class “true” or “fake.” The mechanism is adversarial, so the two networks are optimized alternatively during the training process: the training process is thus designed in such a way that the generator learns both to produce “realistic” samples and to fool the discriminator in its task of detecting the fake samples. The discriminator, in turn, is trained to detect the fake samples and the real ones. Figure 31.6 An overview of how generative adversarial networks work. GANs can be used for image‐to‐image translation [14] or for segmentation problems, in which a setting similar to the image‐to‐image translation has been used [15]. To train a deep neural network, several parameters need to be updated at each training step. While this could be done on a desktop central processing unit (CPU), it would require a massive amount of time. Such a computationally expensive task previously would have required the use of an expensive cluster of CPUs to be run in reasonable amounts of time. As mentioned above, GPUs are commonly used for gaming became they are powerful and cost‐effective, which also makes them suitable for other applications, including medical imaging. The architecture of a GPU is particularly suitable for updating the parameters of a neural network, thanks to the possibility of executing hundreds or thousands of parallel computations, in contrast with the tens that are possible to run on a CPU: for this reason, the training time is reduced by several orders of magnitude. Thanks to the accessibility of powerful gaming GPUs and their relatively low cost, many DL frameworks have been developed, allowing users to train DL models directly using their desktop computers. The most popular software platforms are TensorFlow, Keras, PyTorch, Caffe, and Theano. The majority of frameworks are compatible with the most commonly used programming language for data analysis, such as Python or R, but some frameworks are specific for certain general‐purpose languages such as Java and C++.
31
Artificial Intelligence in Diagnostic Imaging
Introduction
Deep learning
Hardware and Software
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree