(1)
Department of Biology, Trent University, Peterborough, ON, Canada
Abstract
To understand the molecular events responsible for morphological change requires the ability to examine gene expression in a wide range of organisms in addition to model systems to determine how the differences in gene expression correlate with phenotypic differences. There are approximately 12,000 species of butterflies, most, with distinct patterns on their wings. The most important tool for studying gene expression in butterflies is confocal imaging of butterfly tissue by indirect immunofluorescence using either cross-reactive antibodies from closely related species such as Drosophila or developing butterfly-specific antibodies. In this report, we describe how indirect immunofluorescence protocols can be used to visualize protein expression patterns on the butterfly wing imaginal disc and butterfly embryo.
Key words
Butterfly wing imaginal disc stainingConfocal microscopyButterfly embryo stainingIndirect immunofluorescenceLepidoptera1 Introduction
The study of morphological evolution involves the fusion of two distinct disciplines: evolution and developmental biology (evo-devo). Evo-devo seeks to understand how evolutionary change can occur through alterations in gene expression that results in phenotypic alterations. The bulk of the genetic and gene expression data in developmental biology has focused on model systems such as Drosophila melanogaster, Caenorhabditis elegans, and Mus musculus. However, to understand the molecular events responsible for morphological change requires the ability to examine gene expression in a wide range of organisms in addition to model systems to determine how the differences in gene expression correlate with phenotypic differences. The major hurdle facing evo-devo is therefore the ability to develop techniques and methodology to examine gene expression in non-model organisms.
There are approximately 12,000 species of butterflies, most, with distinct patterns on their wings. In fact, even butterflies that are very closely related can have dramatically differing wing patterns [1]. Therefore, butterfly wings represent an important model system to study how patterns are created and how these patterning systems are altered in differing species. Since butterflies represent a non-model system, there are limited tools available to study gene expression.
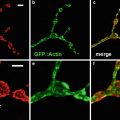
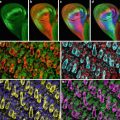
Probably the most important tool for studying gene expression in butterflies is confocal imaging of butterfly tissue by indirect immunofluorescence using either cross-reactive antibodies from closely related species such as Drosophila or developing butterfly-specific antibodies [2–5]. Indirect immunofluorescence protocols from Drosophila have been adapted to work on butterfly tissue [2]; however, a number of differences between Drosophila and butterflies require significant modification of the protocol. First, a thin cellular layer called the peripodial membrane that must be removed prior to confocal imaging of the tissue surrounds butterfly pupal tissue. Secondly, staining of butterfly embryos requires dissection of the embryo out of the egg. Finally, butterfly tissue can be quite large encompassing multiple objective fields on a microscope. Imaging such large tissue samples poses unique challenges when analyzed by confocal microscopy including how to examine expression patterns of genes in large tissue while maximizing the fluorescent signal and retaining a high level of resolution of the tissue. The best method for doing this is to image the large tissue sample by acquiring multiple confocal images using a high power objective. Using readily available computer software such as Adobe Photoshop (Adobe Systems Incorporated, San Jose, CA), the multiple confocal images can be reassembled to generate the entire tissue section to provide a detailed image of the expression pattern of genes throughout the tissue.
To demonstrate how gene expression patterns can be monitored in a non-model system such as butterflies, indirect immunofluorescence will be carried out on butterfly wing discs and embryo tissue using antibodies against Distal-less (Dll) [2], engrailed/invected (en), and cubitus interruptus (Ci) [4].
2 Materials
1.
50 % bleach.
2.
PBS: 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4.
3.
Embryo fix buffer: 0.1 M Pipes, pH 6.9, 1 mM EGTA, pH 6.9, 2 mM MgSO4, 9 % formaldehyde at 4 °C.
4.
Heptane.
5.
Methanol at 4 °C.
6.
Ethanol washes: 25 %, 50 %, 75 % ethanol diluted in water at 4 °C.
7.
Wing disc fix buffer: 0.1 M PIPES, pH 6.9, 1 mM EGTA, pH 6.9, 1 % Triton X-100, 2 mM MgSO4, 1 % formaldehyde.
8.
Block buffer: 50 mM Tris, pH 6.8, 150 mM NaCl, 0.5 % NP40, 5 mg/ml BSA.
9.
Wash buffer: 50 mM Tris, pH 6.8, 150 mM NaCl, 0.5 % NP40, 1 mg/ml BSA.
10.
Vectashield (Vector Laboratories Inc., Burlingame, CA).
11.
Goat anti-rabbit Cy5 (Jackson ImmunoResearch Laboratories, West Grove, PA).
12.
Goat anti-mouse FITC (Jackson ImmunoResearch Laboratories, West Grove, PA).
13.
Goat anti-rat Cy3 (Jackson ImmunoResearch Laboratories, West Grove, PA).
14.
Dissecting microscope.
15.
PBT buffer: 0.1 % Tween 20 in PBS.
3 Methods
The methods outlined below will describe the preparation and fixation of butterfly embryo (1) and wing disc (2) tissue. The tissue will then be stained by indirect immunofluorescence (3) (protocol modified from Brakefield et al., 1996), and the stained tissue will be analyzed using multiple confocal images (4) that will be reassembled to visualize the entire tissue (5).
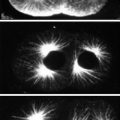
3.1 Butterfly Embryo Fixation Protocol
1.
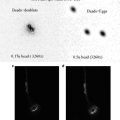
Butterfly eggs are collected at various times after deposition and placed in glass scintillation vials.
2.
The eggs are covered with a 50 % bleach solution and incubated at room temperature for 2 min.
3.
Remove the bleach solution using a Pasteur pipette.
4.
Wash the eggs five times with PBS.
5.
Completely cover the eggs with embryo fix buffer and overlay with a layer of heptane. Incubate at 4 °C for 30 min, shaking occasionally.
6.
Remove the bottom layer of embryo fix buffer, leaving the heptane layer and add 5 ml of methanol. Gently shake the eggs for 15 min.
8.
Rehydrate the eggs by washing in 75 % ethanol followed by a 50 % ethanol wash and finally a 25 % ethanol wash.
9.




Rinse eggs four times in PBS at 4 °C.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree