and Andy Golden1
(1)
Laboratory of Biochemistry and Genetics, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD, USA
Abstract
The microtubule cytoskeleton plays important roles in a number of cellular processes including cell division, establishing and maintaining cell architecture and polarity, and intracellular trafficking. The identification and characterization of factors required for the proper functioning of the microtubule cytoskeleton have been aided by approaches that combine sensitive and rapid methods for high-resolution optical imaging, such as confocal microscopy, with the powerful genetics available in model organisms. Here we present methods for confocal imaging of live and fixed tissues of the nematode C. elegans, a model organism that has been employed with great success to study the microtubule cytoskeleton and its roles in cell division and cell polarity.
Key words
MicrotubulesCentrosomes C. elegans ImmunofluorescenceGFP1 Introduction
Microtubules are linear polymers composed of alpha- and beta-tubulin and possess a defined polarity: a highly dynamic plus end and a less dynamic minus end. Within dividing cells, the microtubule cytoskeleton is in a constant state of flux as it is organized and reorganized to fulfill different purposes. Key to regulating the organization of the microtubule cytoskeleton is the centrosome [1], a non-membrane-bound organelle that nucleates the formation of microtubules such that minus ends are proximal to the centrosome and plus ends distal, leading to the formation of microtubule arrays of uniform polarity. Through the action of microtubule-based motor proteins, these arrays can promote the directed movement of cellular constituents to specific regions of the cell or be organized into a mitotic spindle for the segregation of chromosomes.
Analysis of the behavior and composition of the microtubule cytoskeleton has been aided by the application of confocal microscopy [2]. In combination with a model genetic organism, this imaging technique can be used to study the role of individual genes in controlling various aspects of microtubule behavior. The nematode Caenorhabditis elegans is one such model system that has been used to great advantage to identify genes with microtubule-related functions [3–5]. In the germ cells and embryos of this organism, powerful forward (mutation) and reverse (RNAi) genetic methodologies can be combined with confocal imaging to probe gene function. The large cells of the early embryo have been the preferred tissue for cytological studies. In these cells, robust microtubule arrays, including meiotic and mitotic spindles, are assembled and disassembled according to a rigid developmental plan. This reproducible behavior of the microtubule cytoskeleton allows investigators to readily identify mutations that have even subtle effects on centrosome or microtubule function. Cells of the oocyte and sperm lineages are also amenable to this combined genetic and cytological approach and can be used to study how the microtubule cytoskeleton is specialized to function during oogenesis and spermatogenesis. In this review, we present confocal imaging methods for studying the centrosome and microtubule cytoskeleton in C. elegans embryos and germ cells. Although we present these methods as optimized for the imaging of centrosomes and microtubules, they are applicable to the study of many other intracellular components.
2 Materials
1.
Superfrost slides (Fisher Scientific; Catalog No. 12-550-15).
2.
24 mm × 50 mm cover glasses (Fisher Scientific; Catalog No. 12-545-88).
3.
Tungsten carbide pencil (Fisher Scientific; Catalog No. 13-378).
4.
Poly-l-lysine solution (Sigma; Catalog No. P 8920).
5.
M9 Buffer (1 L) KH2PO4 (3.0 g), Na2HPO4 (6.0 g), NaCl (5.0 g), 1 M MgSO4 (1.0 ml). Mix all but MgSO4. Adjust volume to 999 ml and autoclave. When cooled add sterile 1 M MgSO4.
6.
Anti-α-tubulin monoclonal antibody DM1A (Sigma; Catalog No. T 9026).
7.
10× PBS (1 L) KH2PO4 (2.4 g), Na2HPO4 (14.4 g), NaCl (80.0 g), KCl (2.0 g). Dissolve in 800 ml distilled deionized water. Adjust pH to 7.2, bring volume to 1 L, and autoclave.
8.
PBSBT (0.1 L) Add 10 ml 10× PBS and 0.5 ml Tween 20 to 89.5 ml distilled deionized water. Mix well then add 0.5 g bovine serum albumin (Fraction V). Gently stir the solution until the BSA dissolves. Once made, PBSBT can be stored at 4 °C for 1–2 months.
9.
Alexa Fluorescently labeled secondary antibodies (Molecular Probes).
10.
Vectashield Mounting Medium (Vector Laboratories; Catalog No. H-1000).
11.
TOTO-3 iodide (Molecular Probes; Catalog No. T-3604).
12.
OliGreen (Molecular Probes; Catalog No. O-7582).
13.
37 % Paraformaldehyde (Electron Microscopy Sciences; Catalog No. 15686).
14.
10 % Paraformaldehyde Fixing Solution (10 ml) 37 % paraformaldehyde (2.7 ml), 1 M PIPES, pH 6.7 (1.2 ml). Add components to 6.1 ml water and mix.
15.
16 % Paraformaldehyde (Electron Microscopy Sciences; Catalog No. 15710).
16.
Modified Seydoux and Dunn Fixative (0.1 L) 10× PBS (10 ml), 1 M HEPES pH 6.9 (8 ml), 1 M MgSO4 (0.16 ml), 0.1 M EGTA (0.8 ml), fresh 16 % paraformaldehyde (10 ml). Bring volume to 100 ml with water.
17.
Concavity slides (PGC Scientifics; Catalog No. 60-5500-16).
18.
6 mm × 50 mm disposable glass culture tubes (Fisher Scientific; Catalog No. 14-958-A).
19.
Gonad buffer (1 L) NaCl (3.5 g), KCl (2.4 g), Na2HPO4⋅7H2O (0.8 g), 1 M MgCl2 (2 ml), 1 M CaCl2 (2 ml), 1 M HEPES pH 7.2 (5 ml), 20 % Glucose (10 ml). Add HEPES, NaCl, KCl, and Na2HPO4⋅7H2O to 900 ml water and dissolve thoroughly. Add glucose and mix thoroughly. Add MgCl2 and CaCl2 dropwise to avoid precipitation. Adjust pH if necessary and bring to 1 L with distilled deionized water. Sterilize by filtration.
20.
Silicone grease (Fisher Scientific; Catalog No. 14-635-5D).
21.
Egg buffer (1 L) NaCl (6.9 g), KCl (3.58 g), 1 M MgCl2 (2 ml), 1 M CaCl2 (2 ml), 1 M HEPES pH 7.4 (5 ml). Dissolve NaCl and KCl in 800 ml distilled deionized water then add HEPES, MgCl2, and CaCl2. Adjust pH if necessary and bring to 1 L with water. Autoclave.
22.
Agarose in egg buffer (0.1 L). Add 3 g agarose to 100 ml egg buffer. Heat in a microwave oven until dissolved. Aliquot 4–5 ml per 15 ml disposable polypropylene tube and store at 4 °C.
23.
Lab tape (Daigger; Catalog No. TX14831).
24.
Mouth pipet (Sigma; Catalog No. A-5177).
3 Methods
3.1 Fixation and Staining of Embryos
All protocols for preparing C. elegans for immunofluorescence microscopy include a step to permeabilize the specimen to the fixatives, antibodies, and dyes that are typically used. In intact animals, embryos are protected by a chitinous eggshell and vitelline membrane, and gonads by the cuticle. The eggshell, like the cuticle, is impenetrable by most chemicals and is dealt with in one of two ways. Below we present our version of the popular freeze-crack method in which the eggshell is mechanically disrupted [6, 7]. Embryos are sandwiched between a glass slide and cover glass that is quickly frozen before being snapped apart. There are many permutations of this method, but we find that the protocol below is easy to perform, is highly reproducible, and gives excellent results (Fig. 1a). Alternatively, the eggshell can be removed with alkaline hypochlorite and the embryos stained in suspension [8]. This method, which typically requires a large number of animals, is not suitable for the youngest embryos but does avoid mechanical damage to the specimen.


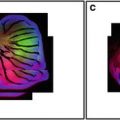
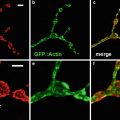
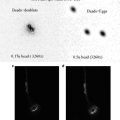
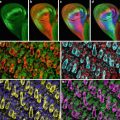
Fig. 1
Confocal images of embryos and germ cells. (a) A two-cell embryo stained for microtubules (green), SPD-2, a centrosome marker (red ), and DNA (blue). The SPD-2 staining appears yellow due to overlap with microtubules. (b) An adult gonad stained with anti-NOP1 antibody (green) to reveal the nucleolus and TOTO-3 (red ) to show DNA. Nuclei in pachytene of meiotic prophase are to the left and oocytes in diakinesis to the right. (c) A primary spermatocyte undergoing meiosis and (d) a field of male germ cells stained for microtubules, SPD-2, and DNA. Bars in a and b, 10 μm. Bars in (c) and (d), 5 μm
Preparation of Slides
1.
Using a tungsten carbide pencil, etch a circle ~15 mm in diameter on the underside of each Superfrost Plus slide to be used (see Note 1). The circle will mark the area of the slide containing the specimen.
2.
Turn the slides over and pipet 10 μl of poly-l-lysine solution onto the surface of the slide (see Note 2).
3.
Using a back-and-forth motion, spread the liquid over the surface with the edge of a glass cover slip. After 5–6 strokes, move to the next slide and do the same. Return repeatedly to each slide re-spreading the liquid until it stays spread and begins to dry.
4.
Leave the slides face up on the bench to finish drying.
Freeze-Cracking
Embryos stain optimally when they are released from the body of the hermaphrodite. This can be accomplished by dissecting gravid hermaphrodites directly on the glass slide (see Note 3).
1.
Draw 18–20 μl of M9 buffer into a micropipette tip.
2.
Dispense approximately two thirds of this in the center of the inscribed circle on the glass slide leaving the rest in the pipet tip.
3.
Using a stereomicroscope, pick hermaphrodites into the drop of M9. To avoid background staining, minimize the amount of bacteria carried over into the M9 buffer. Do not spend more than a few minutes transferring animals as the buffer will evaporate significantly.
4.
Use the edge of a 21-gauge hypodermic needle to cut hermaphrodites in half at the vulva. A well-placed cut will release most of the embryos from the uterus.
5.
Pipet the remaining M9 buffer onto a 24 mm × 50 mm cover glass. Invert the cover glass and turn it so that the long axes of the cover glass and slide are perpendicular. Keeping the cover glass horizontal, gently place it straight down on top of the dissected worms.
6.
Leave the glass sandwich on the bench for 30s to allow the embryos and gonads to adhere to the slide.
7.
Place the glass sandwich on the horizontal surface of a potato masher and gently lower the slide into a Dewar flask of liquid nitrogen (see Note 4). The slide should remain immersed until the liquid nitrogen stops boiling. At this point, the slide can be stored in the liquid nitrogen while you prepare any additional slides.
8.
Using forceps, carefully draw the slides from the liquid nitrogen one by one and immediately remove the cover glass by holding the slide while flicking the overhanging edge of the cover glass with your finger. Immediately proceed to fixation.
Fixation Methods
Methanol Fixation
Methanol is the most common fixative, yields a well-preserved microtubule cytoskeleton, and is compatible with a large number of centrosomal antigens.
Formaldehyde/Methanol Fixation Protocol 1
1.
Immediately after dissecting worms, add an equal volume of 10 % Paraformaldehyde Fixing Solution to the specimen.
2.
Cover specimen with a 24 mm × 50 mm cover glass and leave for 5 min.
Formaldehyde/Methanol Fixation Protocol 2
This fixation protocol was published by Seydoux and Dunn [9] and later modified by J. Schumacher (personal communication). It works well with a wide variety of antibodies.
1.
Immediately after freeze-cracking, submerge slides in –20 °C methanol for 10 s to 1 min.
Staining and Mounting
During the staining process, slides are kept in a humidity chamber to avoid evaporation of antibody solutions during lengthy incubations (see Note 8).
1.




When fixation is complete, transfer the slides to the humidity chamber and pipet 200 μl of blocking solution (PBSBT) onto the specimen. Watch the slide for a minute or so to make sure that the PBSBT stays in contact with the specimen.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree