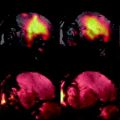
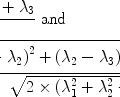Fig. 1.
Flowchart of the CEST imaging protocol for in vivo reporter gene imaging.
2 Materials
2.1 General Requirements
1.
A high-field (4.7 T and above), small animal narrow bore or clinical MR scanner (3 T and above) with a relatively homogeneous main magnetic field, fast and reliable gradient coils, and a high signal-to-noise RF coil.
2.
All the devices for animal anesthesia, motion restraint, physiological monitoring, and body temperature and respiration maintenance are required, for example, an animal holder specifically designed for mouse body imaging with nosepiece adaptor for anesthesia (1–2% isoflurane mixed with air), a physiological (respiration) monitoring and gating system (e.g., SA instruments, Inc., Stony Brook, NY), and an anesthesia induction chamber for use with isoflurane.
3.
For CEST imaging, data post-processing is a crucial step to extract CEST information from the raw images; an image processing software is therefore required and can be chosen from a variety of commercial software packages, including MATLAB (Mathworks, Natick, MA), IDL, etc.
2.2 Gene Cloning, Vector Construction, Cell Culture Conditions, and Transfection
The main advantage of the artificial reporter genes based on CEST is the large variability in the amino acid sequence, since different sequences can provide different CEST contrast.
1.
Lysine-rich protein (LRP) (12) was a prototype reporter gene and a higher level of expression might be achieved due to codon optimization (minimizing repeats) and amino acid sequence optimization (18). The gene can be generated by cloning, in tandem, of DNA oligomers encoding to lysine (AAA or AAG). Two complimentary synthetic oligonucleotides (84 base pairs long), encoding the artificial desired sequence are required, In addition, a start (ATG) codon is required to initiate translation, preferably in the context of a Kozak sequence. Cloning the gene in the frame with an antigen tag (such as HA, V5, myc, etc.) could be useful for subsequent detection with immunohistochemistry.
Alternatively, genes can be synthesized and purchased from biotech companies (e.g., Blue Heron Biotechnology, Bothell, WA).
2.
The expression vectors, either plasmids or viral vectors, should be selected for the specific experiment. The vector should have a promoter that allows the expression of the protein in the right type of cells. For example the cytomegalovirus (CMV) promoter can give expression in most of the mammalian cells but the glial fibrillary acidic protein (GFAP) promoter will drive expression of transgene only in astrocytes in the central nervous system.
3.
For cloning, the following restriction endonucleases are required: BamHI and BglII (New England Biolabs, MA, USA).
4.
Lipofectamine 2000 from Invitrogen is suggested in this chapter; however, different transfection reagents from different manufacturers can be used. Additional reagents and material required for the transfection are Opti-MEM® (reduced serum media, Invitrogen), phosphate-buffered saline (PBS) medium (with serum), and 10-cm culture dishes and 8-well glass chamber slides (Lab-Tek II, Nalgc Nunc, USA). For selection of stably expressing cells, the appropriate selection antibiotics should be used, based on the antibiotic resistance gene carried by the expression vector (e.g., G418, Hygromycin B, Puromycin).
5.
For validation by immunofluorescence the following reagents are needed: a specific antibody that recognizes the fused epitope (e.g., anti-V5 anti-HA, etc.), PBS, cold acetone, Tris-buffered saline Tween-20 (TBST); blocking solution (10% w/v serum in TBST), secondary antibody conjugated to fluorescent dye (e.g., FITC or TRITC) or enzyme (e.g., horseradish peroxidase (HRP) or alkaline phosphatase (AP)).
For protein extraction, the following reagents are required: M-PER® (mammalian protein extraction reagent; Pierce Protein Research Products), dialysis tube (cutoff = 3.5 kDa), protease inhibitor cocktail (e.g., Halt™; Pierce Protein Research Products), and Bradford assay (Pierce Protein Research Products).
2.3 MRI
The following pulse sequences should be available.
1.
A fast 3D localizer sequence and sequences for anatomic imaging, such as multi-slice spin echo (SE) and multi-slice gradient echo (GRE).
2.
A localized spectroscopy sequence, such as PRESS (point-resolved spectroscopy).
3.
A fast spin echo sequence, such as RARE (rapid acquisition with relaxation enhancement), or a fast gradient echo sequence, such as EPI (echo planar imaging), including a magnetization transfer (MT) module, with which a saturation pulse can be manipulated with respect to the desired pulse shape, power, duration, and offset. The MRI scanner should be equipped with an isoflurane inhalation setup and animal cradle.
2.4 General Lab Equipment
Humidified incubator set at 37°C , benchtop centrifuges, upright and inverted microscopes.
2.5 B0 Correcting Algorithm
The analytic expression for the steady state saturation lineshape for the longitudinal magnetization 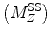 of a single proton, is given by (22):
of a single proton, is given by (22):
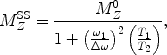 where
where 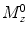 is the initial longitudinal magnetization, ω 1 is the strength of the applied RF pulse, which resonates at Δω (frequency with respect to the proton center frequency). This formula is implemented into a MATLAB algorithm (see Section 3.5).
is the initial longitudinal magnetization, ω 1 is the strength of the applied RF pulse, which resonates at Δω (frequency with respect to the proton center frequency). This formula is implemented into a MATLAB algorithm (see Section 3.5).
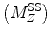 of a single proton, is given by (22):
of a single proton, is given by (22):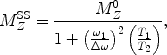
 is the initial longitudinal magnetization, ω 1 is the strength of the applied RF pulse, which resonates at Δω (frequency with respect to the proton center frequency). This formula is implemented into a MATLAB algorithm (see Section 3.5).
is the initial longitudinal magnetization, ω 1 is the strength of the applied RF pulse, which resonates at Δω (frequency with respect to the proton center frequency). This formula is implemented into a MATLAB algorithm (see Section 3.5).3 Methods
3.1 Cloning and Transfection
3.1.1 Cloning of CEST Reporter Gene
Two complimentary synthetic oligonucleotides (84 base pairs long), encoding the artificial desired sequence, should be designed, so that, after annealing, they will retain corresponding endonuclease restriction site overhangs (e.g., to BglII at the 5′-end and BamHI at the 3′-end). This double-strand DNA is referred to as the monomer. The monomer should be cloned into the expression vector in the context of the desired promoter that will allow expression in the target cells. After cloning, the new vector should be digested with BglII and BamHI, and the released insert should be ligated into the BglII sites of the parental vector (See Note 1). The ligation will result in a dimer (i.e., a sequence encoding to polypeptide with double length). Next, the new vector should be digested with BglII and BamHI and the released insert (dimer) should be ligated into the BglII sites of the parental vector to form a tetramer. This process should be repeated until the DNA sequence is sufficiently long (see Note 2).
3.1.2 Cell Culture and Transfection
The cell line of choice can be transfected with the new construct. In general, any transfection or viral infection protocol is adequate; here, we briefly describe a general protocol for transfection using Lipofectamine 2000 from Invitrogen (See Note 3).
A day before transfection, the cells should be plated in 10-cm tissue culture dish in a dilution such that the cells will be at 80% confluence on the transfection day.
1.
Lipofectamine 2000 (60 μl) should be diluted in 1 mL optiMEM, mixed gently, and allowed to stand for 5 min. Then, 24 μg DNA should be added to 1 mL optiMEM. The diluted DNA and lipofectamine solution should be combined and incubated for 20 min at room temperature.
Next, the cells should be washed with optiMEM or PBS and the medium should be replaced with 8 mL optiMEM.
2.
The DNA transfection mixture (2 mL) should then be added gently to each well and incubated at 37°C for 5–6 h.
3.
Then, regular medium (with serum) should be used.
4.
In order to generate stable clones, the media should be supplemented with the appropriate selection of antibiotics (e.g., G418, hygromycin B, puromycin).
3.1.3 Validation by Immunofluorescence
In order to validate the protein expression, the cells could be stained using a specific antibody that recognizes the fused epitope (e.g., anti-V5 anti-HA, etc.). The protocol below can be modified and optimized depending on the antibodies and cell (see Note 4).
The cells can be grown on 8-well glass chamber slides (Lab-Tek II, Nalge Nunc, USA) overnight. The volumes may need to be adjusted for different plates.
1.
The cell culture medium can then be removed, the cells washed with PBS, and fixed with cold acetone for 10 min at –20°C.
2.
After air-drying for 15 min, the cells should be washed twice, for 5 min each time, with TBST (0.2 mL/wash).
3.
Next, 0.1 mL of blocking solution (PBS containing serum) should be added and the mixture should be incubated for 60 min at room temperature to reduce non-specific binding of antibody.
4.
The blocking solution should be removed and 0.1 mL of PBS should be added (containing serum), with the appropriate first antibody (1:100 to 1:400 dilution of antibody). This should be incubated for 60 min at room temperature or overnight at 4°C.
5.
The cells should be washed three times, for 5 min each time, with PBS and then incubated in 0.1 mL of PBS containing the appropriate secondary antibody conjugated to fluorescent dye or enzyme for 60 min at room temperature in the dark.
6.
Then, wash cells three times, for 5 min each time, with PBS again, and continue counter-staining.
7.
Finally, the cells should be mounted with mounting media, covered with a cover-slip, and the results should be analyzed with microscopy.
3.1.4 Protein Extraction
As CEST reporter genes are extremely sensitive, the changes in pH and the exchange rate are dramatically reduced when the cells are fixed. The best way to measure the CEST contrast in vitro is to extract the proteins. The cells should be washed twice and collected in 10 mM of ice-cold PBS (pH = 7.1, without Mg2+/Ca2+). After centrifugation at 2,500g for 10 min at 4°C, the pellet should be suspended in 1 mL of M-PER® (mammalian protein extraction reagent Pierce Protein Research Products) and shaken gently for 10 min at 4°C. Cell debris should be removed by centrifugation at 14,000g for 15 min at 4°C. The supernatant should be transferred to a dialysis tube (cutoff = 3.5 kDa) and dialyzed twice against PBS. A protease inhibitor cocktail should be added (e.g., Pierce Protein Research Products; Halt™) and the protein extraction should be stored at –80°C. Protein concentrations can be determined using the Bradford assay (Pierce Protein Research Products).
3.1.5 Transplantation
Animal procedures should be conducted in accordance with the guidelines for the care and use of research animals. The cells should be collected and suspended in a low volume of PBS or saline. The cells should be inculcated into the target tissue while the animal is kept anesthetized by isoflurane inhalation (1–2%).
3.2 General MRI Protocol
3.2.1 Localizer
1.




In vivo MR imaging should be performed several days after cell transplantation. The animals should be restrained in an animal cradle, centered at both the center of the RF coil and the center of the magnet, and kept anesthetized using 1–2% isoflurane gas throughout the imaging procedure.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree